私立中山醫學院醫學研究所碩士論文
Master Thesis, Institute of Medicine, Chung Shan Medical and Dental College
對金黃色葡萄球菌上抗鎘腺核甘三磷酸酶穿膜
結構之研究
Membrane Topology of CadA Cadmium
Resistant ATPase in Staphylococcus
aureus
指導教授 : 蔡淦仁 (Tsai, Kan-Jen) 博士
研究生 : 林詠峰 (Lin, Yung-Feng) 撰
博碩士論文電子檔案上網授權書
本授權書所授權之論文為授權人在中山醫學院醫學研究所 89 學年度 第二學期取得碩士學位之論文。 論文題目:對金黃色葡萄球菌上抗鎘腺核甘三磷酸酶穿膜結構之研究 指導教授:蔡淦仁 博士 茲同意將授權人擁有著作權之上列論文全文(含摘要),非專屬、無償 授權國家圖書館及授權人畢業學校之圖書館,不限地域、時間與次 數,以微縮、光碟或其他各種數位化方式將上列論文重製,並得將數 位化之上列論文及論文電子檔以上載網路方式,提供讀者基於個人非 營利性質之線上檢索、閱覽、下載或列印。 ● 讀者基於非營利性質之線上檢索、閱覽、下載或列印上列論文,應依著作權 法相關規定辦理。授權人
簽 名:
林詠峰
中 華 民 國 90 年 6 月 28 日本論文為中山醫學院授與理學碩士學位之必備條件之一,經中山醫學 院醫學研究碩士論文考試委員會審查合格及口試通過。
口試委員
中國醫藥學院微生物研究所副教授 劉昭君 博士 ___________ 中山醫學院毒理學研究所教授 林嬪嬪 博士 ___________ 中山醫學院醫學研究所副教授 蔡淦仁 博士 (論文指導教授) ___________中華民國九十年六月
學生林詠峰論文題目為對金黃色葡萄球菌上抗鎘腺核甘三磷酸酶穿 膜結構之研究,其論文已經私立中山醫學院醫學研究所論文碩士委員 審查合格及口試通過,並由其指導教授校閱後無誤。
中華民國九十年六月二十二日
誌 謝
本篇論文的完成要感謝很多人的幫忙。除了感謝母校中山提供 的研究機會之外,首要感謝的是我論文指導教授蔡淦仁老師。在這兩 年期間,他教導我如何從一個學生變成一位學者。幽默風趣的教學方 法,理性尊重的待人風格,讓我著實受益良多。再者感謝林嬪嬪老師 及劉昭君老師,在我邁向碩士學位的最後關頭助我一臂之力。另外也 要感謝美國Rosen 博士的實驗室提供了重要的實驗材料和許多的建 議,以及日本Nakae 博士的實驗室贈送了實驗用的質體。還有我們實 驗室的淑娟、宏基和佳瑜,已畢業的學樑和秉儒,隔壁實驗室的澤民 以及美國朋友范斌和Marco 提供了實驗上的協助。也感謝醫研所和 醫技系的老師們,整個大實驗室裏的所有夥伴們以及其他不勝列舉的 朋友們,提供有形和無形的幫助。最後謹將這份小小的成就和喜悅獻 給我的母親詹氏和夫人惠雯以及我親愛的家人們。 研究生 林詠峰 謹述于 中山醫學院研究大樓十三樓實驗室中華民國九十年六月 摘 要 細菌對於有毒金屬之抗性起初是來自於自然環境裏的暴露,而最近則 被發現有來自於藥品的使用。有一些細菌的抗性已知是由添加性的突 變所產生的。之前的研究發現有一盤尼西林(penicillin)抗性質體 pI258 提供了金黃色葡萄球菌(Staphylococcus aureus)對鎘離子的抗性。生化 學上對於該種抗性的探討則已發現這是由pI258 上 cadA 基因所轉錄 的CadA 抗鎘腺核甘三磷酸酶(Cd2+-ATPase)所提供的抗性。序列比對
的結果,將此CadA 蛋白歸類於 P 型腺核甘三磷酸酶(P-type ATPase)。
另因CadA 這類蛋白在結構上有另外的特徵,該蛋白和其它一些能轉 運重金屬離子的腺核甘三磷酸酶則進一步被歸類在一新的CPx 型腺 核甘三磷酸酶(CPx-type ATPase)次族群裏。在這次的研究當中,我們 已經探查出CadA 蛋白的穿膜結構以及 CPx 型腺核甘三磷酸酶這一類 蛋白所特有的一些功能性區域所在的位置。利用鹼性磷酸酶(phoA)和 乙型半乳糖酶(lacZ)基因融合的方法,我們製造出各二十二種的 cadA-phoA 和 cadA-lacZ 的基因融合,涵蓋了該轉運蛋白的全長。然 後根據這些融合酵素活性的高低,我們分析後得到的結果是 CadA 蛋 白擁有八個穿膜片段(TMs),其中第三和第四片段可能不完全穿膜; 氮端(N)和碳端(C)都留在細胞質中。這樣的結果跟另外一個發現於幽 門桿菌(Helicobacter pylori)的 CadA 同源蛋白的穿膜結構研究的結果 很相近,除了對第三和第四穿膜片段的描述之外。另外則有一些結構 上的差異被發現存在於這兩個同源蛋白之間,包括葡萄球菌上的 CadA 含有一高厭水性的區域緊接在假設的腺核甘三磷酸結合位置 (ATP binding site)後面,以及在這兩個蛋白之間其帶電胺基酸的分佈 也是不相稱的。總而言之, 以葡萄球菌上的 CadA 當一個例子, 我們
Abstract
Bacterial metal resistances evolved originally from the exposure
of natural sources, and become more severe in recent years after
the usages of therapeutic drugs. Some of these bacterial resistant
mechanisms are raised from additive mutations of bacterial
genes. Previously, the staphylococcal resistance to cadmium
carried by pI258 penicillinase plasmid has been extensively
studied. Biochemical characterization of the resistance has
revealed that a CadA Cd
2+-ATPase mediates the resistance
phenomena and this protein has been categorized into a P-type
ATPase enzyme family. Due to their difference in certain
structural features, CadA and other P-type ATPases for
transporting heavy metal ions have been grouped into a new
enzyme family, called CPx-type ATPases. In this proposed study,
we have investigated the CadA topography and, as a model, to
characterize those functional domains unique for the
CPx-ATPases. Using the phoA (alkaline phosphatase) and lacZ
(β-galactosidase) gene fusion methods, a series of 22 cadA-phoA
and 22 cadA-lacZ fusions were created throughout the cadA
gene. Based on the reporter enzyme activities measured in this
study, we have determined that there are eight transmembrane
segments (TMs) in CadA; however, the third and forth segments
might not traverse the membrane completely. Both the
N-terminus and the C-terminus of CadA protein are located
within the cytoplasm. This topological model is similar to that of
the homologous ATPase found in Helicobacter pylori except the
description of the TM3 and TM4. Besides, some significant
differences have been found between these two pumps,
including a highly hydrophobic region just behind the putative
ATP binding site found in staphylococcal CadA but not in the
helicobacter version of CadA, and an asymmetric arrangement
of charged residues between these two ATPases. Anyway, we
proposed a more detailed structure model of this kind of
ATPases using staphylococcal CadA as an example.
Institute of Medicine, Chung Shan Medical and Dental College
Membrane Topology of CadA Cadmium
Resistant ATPase in Staphylococcus aureus
Yung-Feng Lin
June, 2001
Table of Contents
Table of Contents ………. i
List of Tables ……… iii
List of Figures ……….. iv
Chapter 1. Introduction ……….………… 1
1.1 Bacterial resistance to toxic metals ……… 1
1.1.1 Cadmium resistance ………. 1
1.1.2 CadA is a cadmium efflux in Staphylococcus aureus ….. 3
1.1.3 The cadA operon in S. aureus pI258 ……… 4
1.2 CadA is a metal transporting P-type ATPase ………. 5
1.2.1 CadA is a member of P-type ATPases ………. 6
1.2.2 The heavy metal transporting CPx-type ATPase ………. 9
1.3 The topological study of CPx-type ATPases ….……….. 11
1.3.1 Inferences from sequence- hydropathy analysis ….…….. 11
1.3.2 Experimental approaches to membrane topology ……... 13
1.3.3 Previous membrane topology studies in CPx-type ATPase related proteins ..…….……….……… 18
1.4 Motivations for CadA topology ……….………. 20
1.4.1 Gene fusion strategy for CadA topology ……….………. 21
1.4.2 Properties of reporter enzymes ……….……… 23
1.5 Rationale and experimental approach ……….……… 23
Chapter 2. Materials and Methods ……….……... 25
2.1 Bacterial strains, plasmids, and culture conditions …….……… 25
ii
2.3 Construction of inducible cadA expression system ……… 27
2.4 Cadmium resistance assays ……… 27
2.5 Construction of cadA-phoA and cadA-lacZ fusions …………... 29
2.5.1 CadA-phoA fusion ……….. 29
2.5.2 CadA-lacZ fusion ……… 31
2.6 DNA Sequencing ………... 31
2.7 Expression of the hybrid proteins ……….. 31
2.8 Enzyme activity assays ……….. 32
2.8.1 Alkaline phosphatase ……….. 32
2.8.2 β-galactosidase ……… 33
2.9 Cell fractionation ……… 33
2.10 Protein electrophoresis and Western blotting ………... 34
Chapter 3. Results ……….. 36
3.1 Determine the cadmium resistance of cells harboring pKJ100 plasmid ………... 36
3.2 Computer hydropathy analysis ………... 36
3.3 Isolation of cadA-phoA and cadA-lacZ fusions ……….. 38
3.4 Determination of the first transmembrane segment ……… 39
3.5 Enzyme activity analysis of cadA-phoA and cadA-lacZ fusions . 42 3.6 Determine the protein productions of CadA fusion ……… 44
Chapter 4. Discussion ………. 49
List of Tables
Table I Strains and plasmids ……… 26 Table II Primers ………... 30 Table III Nucleotide sequence of the cadA-phoA and cadA-lacZ
fusion junction ……… 40 Table IV Fusion enzyme activities ……… 43 Table V Molecular weights of the fusion proteins ……….. 48
iv
List of Figures
Fig. 1. The schematic model of CPx-type ATPases ……….. 8 Fig. 2. Hydropathy plots ……… 14 Fig. 3. Use of fusions to analyze membrane protein topology …….…. 17 Fig. 4. Comparison of S. aureus CadA and the homologous H. polori
ATPase ……… 19 Fig. 5. Constructed plasmids ………. 28 Fig. 6. Metal resistance of CadA ……….….. 37 Fig. 7. Determination of the first transmembrane segment of CadA by
western blotting ……….….. 41 Fig. 8. Topological structure of the CadA protein ……….…… 45 Fig. 9. Western blots of cell extracts of various fusions …………..….. 47
Chapter 1. Introduction
1.1 Bacterial resistance to toxic metals
Bacterial resistance to toxic substances is ubiquitously found in the
contaminated environment (Foye 1977, Knowles et al. 1983). For their survivals in the presence of toxic metal ions, various mechanisms are developed to confer resistance to these environmental toxins, including those resistances to arsenate, arsenite, antimony, lead, bismuth, zinc, mercury, as well as cadmium (Novick and Roth 1968, Foster 1983). Some of these resistance mechanisms are adaptive from additive mutations of some existed bacterial genes, and some are from alternative substrate usages of the bacterial genes (Aguiar et al., 1990; Inbar and Ron, 1993; Hustavova et al., 1995; LaRossa et al., 1995; Crupper et al., 1999). And the resistance genes spread among closely related species and even to other species through transformation, transduction and transposition in the bacterial world. Particularly, plasmid-mediated resistances to metal ions are frequently found in prokaryotes (Rensing et al., 1999).
1.1.1 Cadmium resistance
Cadmium is a hazard element universally found on earth, and this metal ion is one of the most likely contaminants found during the industry evolution. Bacteria and some other microorganisms have
developed certain mechanisms to cop with the cadmium toxicity (Aguiar
et al., 1990; Inbar and Ron, 1993; Hustavova et al., 1995; LaRossa et al.,
私立中山醫學院醫學研究所碩士論文
Master Thesis, Institute of Medicine, Chung Shan Medical and Dental College
對金黃色葡萄球菌上抗鎘腺核甘三磷酸酶穿膜
結構之研究
Membrane Topology of CadA Cadmium
Resistant ATPase in Staphylococcus
aureus
指導教授 : 蔡淦仁 (Tsai, Kan-Jen) 博士
研究生 : 林詠峰 (Lin, Yung-Feng) 撰
博碩士論文電子檔案上網授權書
本授權書所授權之論文為授權人在中山醫學院醫學研究所 89 學年度 第二學期取得碩士學位之論文。 論文題目:對金黃色葡萄球菌上抗鎘腺核甘三磷酸酶穿膜結構之研究 指導教授:蔡淦仁 博士 茲同意將授權人擁有著作權之上列論文全文(含摘要),非專屬、無償 授權國家圖書館及授權人畢業學校之圖書館,不限地域、時間與次 數,以微縮、光碟或其他各種數位化方式將上列論文重製,並得將數 位化之上列論文及論文電子檔以上載網路方式,提供讀者基於個人非 營利性質之線上檢索、閱覽、下載或列印。 ● 讀者基於非營利性質之線上檢索、閱覽、下載或列印上列論文,應依著作權 法相關規定辦理。授權人
簽 名:
林詠峰
中 華 民 國 90 年 6 月 28 日本論文為中山醫學院授與理學碩士學位之必備條件之一,經中山醫學 院醫學研究碩士論文考試委員會審查合格及口試通過。
口試委員
中國醫藥學院微生物研究所副教授 劉昭君 博士 ___________ 中山醫學院毒理學研究所教授 林嬪嬪 博士 ___________ 中山醫學院醫學研究所副教授 蔡淦仁 博士 (論文指導教授) ___________中華民國九十年六月
學生林詠峰論文題目為對金黃色葡萄球菌上抗鎘腺核甘三磷酸酶穿 膜結構之研究,其論文已經私立中山醫學院醫學研究所論文碩士委員 審查合格及口試通過,並由其指導教授校閱後無誤。
中華民國九十年六月二十二日
誌 謝
本篇論文的完成要感謝很多人的幫忙。除了感謝母校中山提供 的研究機會之外,首要感謝的是我論文指導教授蔡淦仁老師。在這兩 年期間,他教導我如何從一個學生變成一位學者。幽默風趣的教學方 法,理性尊重的待人風格,讓我著實受益良多。再者感謝林嬪嬪老師 及劉昭君老師,在我邁向碩士學位的最後關頭助我一臂之力。另外也 要感謝美國Rosen 博士的實驗室提供了重要的實驗材料和許多的建 議,以及日本Nakae 博士的實驗室贈送了實驗用的質體。還有我們實 驗室的淑娟、宏基和佳瑜,已畢業的學樑和秉儒,隔壁實驗室的澤民 以及美國朋友范斌和Marco 提供了實驗上的協助。也感謝醫研所和 醫技系的老師們,整個大實驗室裏的所有夥伴們以及其他不勝列舉的 朋友們,提供有形和無形的幫助。最後謹將這份小小的成就和喜悅獻 給我的母親詹氏和夫人惠雯以及我親愛的家人們。 研究生 林詠峰 謹述于 中山醫學院研究大樓十三樓實驗室中華民國九十年六月 摘 要 細菌對於有毒金屬之抗性起初是來自於自然環境裏的暴露,而最近則 被發現有來自於藥品的使用。有一些細菌的抗性已知是由添加性的突 變所產生的。之前的研究發現有一盤尼西林(penicillin)抗性質體 pI258 提供了金黃色葡萄球菌(Staphylococcus aureus)對鎘離子的抗性。生化 學上對於該種抗性的探討則已發現這是由pI258 上 cadA 基因所轉錄 的CadA 抗鎘腺核甘三磷酸酶(Cd2+-ATPase)所提供的抗性。序列比對
的結果,將此CadA 蛋白歸類於 P 型腺核甘三磷酸酶(P-type ATPase)。
另因CadA 這類蛋白在結構上有另外的特徵,該蛋白和其它一些能轉 運重金屬離子的腺核甘三磷酸酶則進一步被歸類在一新的CPx 型腺 核甘三磷酸酶(CPx-type ATPase)次族群裏。在這次的研究當中,我們 已經探查出CadA 蛋白的穿膜結構以及 CPx 型腺核甘三磷酸酶這一類 蛋白所特有的一些功能性區域所在的位置。利用鹼性磷酸酶(phoA)和 乙型半乳糖酶(lacZ)基因融合的方法,我們製造出各二十二種的 cadA-phoA 和 cadA-lacZ 的基因融合,涵蓋了該轉運蛋白的全長。然 後根據這些融合酵素活性的高低,我們分析後得到的結果是 CadA 蛋 白擁有八個穿膜片段(TMs),其中第三和第四片段可能不完全穿膜; 氮端(N)和碳端(C)都留在細胞質中。這樣的結果跟另外一個發現於幽 門桿菌(Helicobacter pylori)的 CadA 同源蛋白的穿膜結構研究的結果 很相近,除了對第三和第四穿膜片段的描述之外。另外則有一些結構 上的差異被發現存在於這兩個同源蛋白之間,包括葡萄球菌上的 CadA 含有一高厭水性的區域緊接在假設的腺核甘三磷酸結合位置 (ATP binding site)後面,以及在這兩個蛋白之間其帶電胺基酸的分佈 也是不相稱的。總而言之, 以葡萄球菌上的 CadA 當一個例子, 我們
Abstract
Bacterial metal resistances evolved originally from the exposure
of natural sources, and become more severe in recent years after
the usages of therapeutic drugs. Some of these bacterial resistant
mechanisms are raised from additive mutations of bacterial
genes. Previously, the staphylococcal resistance to cadmium
carried by pI258 penicillinase plasmid has been extensively
studied. Biochemical characterization of the resistance has
revealed that a CadA Cd
2+-ATPase mediates the resistance
phenomena and this protein has been categorized into a P-type
ATPase enzyme family. Due to their difference in certain
structural features, CadA and other P-type ATPases for
transporting heavy metal ions have been grouped into a new
enzyme family, called CPx-type ATPases. In this proposed study,
we have investigated the CadA topography and, as a model, to
characterize those functional domains unique for the
CPx-ATPases. Using the phoA (alkaline phosphatase) and lacZ
(β-galactosidase) gene fusion methods, a series of 22 cadA-phoA
and 22 cadA-lacZ fusions were created throughout the cadA
gene. Based on the reporter enzyme activities measured in this
study, we have determined that there are eight transmembrane
segments (TMs) in CadA; however, the third and forth segments
might not traverse the membrane completely. Both the
N-terminus and the C-terminus of CadA protein are located
within the cytoplasm. This topological model is similar to that of
the homologous ATPase found in Helicobacter pylori except the
description of the TM3 and TM4. Besides, some significant
differences have been found between these two pumps,
including a highly hydrophobic region just behind the putative
ATP binding site found in staphylococcal CadA but not in the
helicobacter version of CadA, and an asymmetric arrangement
of charged residues between these two ATPases. Anyway, we
proposed a more detailed structure model of this kind of
ATPases using staphylococcal CadA as an example.
Institute of Medicine, Chung Shan Medical and Dental College
Membrane Topology of CadA Cadmium
Resistant ATPase in Staphylococcus aureus
Yung-Feng Lin
June, 2001
resistance systems to cadmium have been reported to date. For example, Nies et al. (1987) isolated a 9.1-kb EcoRI fragment from a 238-kb
plasmid, pMOL30, found in Alcligenes eutrophus CH34, which confers resistance to cadmium, zinc and cobalt salts in sensitive strains. Later, Nies et al. has also reported that the genes contribute to these three metals resistance arranged as a czc operon, which encodes five polypeptides called CzcA, B, C, D, and R. The CzcR and Czc D are regulatory
elements for the expression of CzcA, B, and C, the structure genes of the resistance system, in which the CzcA, CzcB and CzcC are essential for a full resistance to all three metals cations (Nies and Silver, 1989; Nies et
al., 1989; Nies, 1992). Similarly, some cadmium resistance systems were
found in Staphylococci spp. In S. aureus, two resistant determinants to cadmium were identified in staphylococcal plasmid pI258, called cadA and cadB (Novick and Roth 1968; Smith and Novick, 1972; Weiss et al., 1978; Novick et al., 1979). Recently, Chaouni et al. (1996) reported a third cadmium resistant system exists in pLUG10 plasmid from S.
lugdunensis. In this latter resistant system, two genes were identified,
which includes a cadB-like cadmium resistance gene and a regulatory gene, called cadX, which functions in a high-level of cadmium resistance.
Studies have shown that the cadA and cadB resistant determinants are different in their resistance mechanisms even though they are both carried by plasmid pI258 in S. aureus (Novick and Roth 1968; Smith and Novick, 1972; Weiss et al., 1978; Novick et al., 1979). It was suggested that the cadA resistant determinant is mediated by a 727-amino-acid protein, which shows sequence similarity to the P-type ATPase (Nucifora
et al., 1989). On the other hand, the cadmium resistance mediated by the cadB resistant determinant was poorly defined. However, it has been
3
reported that cadmium resistance of the cadB resistant determinant does not due to the decrease of cadmium accumulation within the resistant cells (Smith and Novick, 1972; Perry and Silver, 1982). It was proposed that CadB does not promote cation efflux as that of CadA, the latter system was characterized as an efflux pump for the cadmium resistance, and the CadB may provide a protection through the binding of cadmium on the membrane (Perry and Silver, 1982). Another cadmium resistance gene found in S. aureus, cadD, has also been recently identified from the plasmid pRW001 (Crupper et al., 1999). A high degree of sequence similarity was found between cadD and the cadB-like gene from S.
lugdunensis; however, no evidence related to either cadA or cadB was
found. The expression of cadD in S. aureus as well as Bacillus subtilis has shown a low-level resistance to cadmium when compared it with that of cadA and cadB (Crupper et al., 1999). Suggesting that the resistance to cadmium mediated by cadD gene plays a minor role and only functions when other cadmium resistant determinants were absence.
1.1.2 CadA is a cadmium efflux in Staphylococcus aureus
The cadA gene from staphylococcal penicillinase plasmid pI258 was
identified along with many other resistant determinants to heavy metal ions in S. aureus (Novick and Roth, 1968). Early study from Chopra (1975) has shown that protein synthesis in cell-free extracts from resistance or sensitive bacteria was equally susceptible to inhibition by Cd2+; however, the spheroplasts prepared from resistant bacteria retained their resistance. Based on these findings, a cadmium resistance due to the change in the orientation of the membrane protein or phospholipids was
proposed. The Cd2+ enters into cells via an energy-dependent Mn2+
transport system as an alternative substrate for the transport system in S.
aureus and maybe same to other Gram-positive bacteria (Tynecka et al.,
1981). Similarly, Weiss et al. (1978) demonstrated that Cd2+ inhibits the uptake of Mn2+ and accelerates the loss of intracellular of Mn2+ in the susceptible cells, but no effect was found on Mn2+ transport in resistant S.
aureus. Weiss et al. (1978) has also suggested that the cadA resistance
mechanism is dependent upon a blockage of energy-dependent Cd2+ transport. On the other hand, Tynecka et al. (1981) found that the resistance was mediated by an active efflux pump to reduce the
intracellular accumulation of the toxic metal ions and the net efflux of Cd2+ was blocked in the presence of 2,4-dinitrophenol, N, N,
-dicylohexylcarbodiimide (DCCD) or incubation at 4˚C. Indicating that the resistance is in an energy-dependent fashion (Tynecka et al., 1981). Furthermore, they also found that valinomycin had no effect on the cadmium resistance and a Cd2+/2H+ antiporter for cadmium resistance was then proposed (Tynecka et al., 1981).
1.1.3 The cadA operon in S. aureus pI258
The most striking finding in cadA cadmium resistant determinant was come from the identification of the genes involved in the resistance (Nucifora et al. 1989). Two open reading frames (ORFs) were found within the cadmium resistant determinant, called cadC and cadA genes, and later the cadmium determinant was named cad operon. In their report, they found that a 3.5-kb BglII-XbaI DNA fragment from plasmid pI258 contains the cadA operon to confer a full cadmium resistance, however,
5
the large ORF, the cadA gene, seemed more essential for the resistance phenomena (Nucifora et al. 1989). When cadA and cadC were both expressed in S. aureus, the full complementation of cadmium resistance was observed when compared it with that of cadA or cadC gene alone (Yoon and Silver, 1991).
When aligned the deduced amino acid sequence of cadA gene with the sequences from other known proteins, it was found that the predicted CadA protein shows a high homology to a class of enzymes called E1E2-ATPase or P-type ATPase (Nucifora et al., 1989; Silver et al.,
1989). On the other hand, the smaller ORF with a 3’ end of the gene overlaps 8 base pairs to cadA encodes for a short polypeptide of 122 amino acids in length was named cadC (Nucifora et al., 1989).
Furthermore, it was found that purified CadC protein could bind to the proposed cadA operator/promoter DNA sequences, and Cd2+, Bi3+ and Pb2+ caused the releases of CadCs from DNA in gel retardation assays and DNaseI footprinting experiments (Endo and Silver, 1995). These observation strongly suggested that cadC is a regulatory element in cad operon, as it was previously found that the predicted cadC gene product shown homology only to the regulatory element of Ars operon, the ArsR gene (Wu and Rosen, 1991). A regulatory role for cadC gene was further confirmed by co-transformed this gene with a report gene under
controlled by the cadA-operator/promoter in E. coli (Rensing et al., 1998). Therefore, the regulatory role of cadC to modulate the cadA expression in the cad operon in S. aureus should be without any suspicion.
1.2.1 CadA is a member of P-type ATPases
As mentioned in the above section, the amino acid sequences of predicted cadA gene product is similar to other P-type ATPase
transporters (Nucifora et al., 1989; Silver et al., 1989; Silver and Walderhaug, 1992). The cadA gene was cloned and expressed it in
Bacillus substilis (Nucifora et al., 1989). In vitro transport study has
confirmed that CadA mediates an ATP-dependent cadmium efflux to confer resistance to cadmium (Tsai et al., 1992). Using everted membrane vesicles prepared from B. subtilis harboring cadA gene and in the
presence of different energy sources in the cadmium transport assay. They showed that the cadmium transport was driven only in the presence of ATP, but not NADH or phenazine methosulfate plus sodium ascorbate, which together generate a proton motive force. They also showed that bafilomycin A1 inhibited to cadmium transport at a concentration of
micro molar range, which is comparable to other P-type ATPases. Furthermore, a cadmium-dependent CadA protein phosphorylated
intermediate was demonstrated within the CadA enzyme cycle (Tsai and Linet, 1993). Together all these evidence, the role of CadA as a
cation-translocating ATPase similar to those of P-type ATPases was established.
The P-type ATPases are polytopic membrane proteins and their major functions are transporting cations against their concentration gradients with the expenses of chemical energy, ATP. Characteristically, these ATPases form a covalently phosphorylated intermediate in their reaction cycles (Pedersen and Carafoli, 1987a and 1987b). Most P-type ATPases are single, large, and catalytic monomers ranging from 70 to 200 kDa in
7
size, with an exception of Na+/K+-ATPase found in eukaryotes, in which heterodimer mediated the transport activity, and this type of ATPases are inhibited by micro molar concentrations of vanadate. Additionally, all P-type ATPases share a unique characteristic that is the formation of phosphorylated aspartate, which is formed transiently when the energy from ATP hydrolyzation is used to translocate the substrate, as the hallmark of this enzyme family. More recently, it was found that the invariant aspartic acid (D) residue in the conserved sequence DKTGT is first phosphorylated while ATP binds to the ATP-binding domain (Solioz and Vulpe, 1996, Fig.1). The conserved sequences found in these
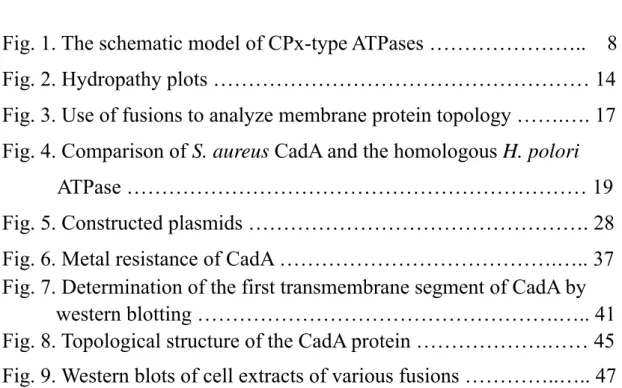
ATPases include the kinase domain (DKTGT) as described, an ATP binding domain (GDGXNDXP) and a phosphatase domain (TGES/A) (Solioz and Vulpe, 1996; Fig. 1).
The best-known P-type ATPases include eukaryotic plasma
membrane Na+/ K+-ATPase, sarcoplasmic reticulum Ca2+-ATPase (Shull
et al., 1985; MacLennan et al., 1985), yeast plasma membrane Na+/K+-, K+- and Ca2+-like ATPases (Serrano et al., 1986), and bacterial
K+-ATPase from E. coli. More recently, some newly found eukaryotic and prokaryotic ATPases were also categorized into this enzyme family,
which include the iron-induced mammalian iron ATPase (Baranano et al., 2000), the facultatively anaerobic alkaliphile Na+-ATPase from
Exiguobacterium aurantiacum (Ueno et al., 2000), and a soluble P-type
ATPase from Methanococcus jannaschii (Ogawa et al., 2000). In addition to these essential metal-translocating pumps, P-type ATPases were also found in the transporting of heavy metal ions, such as CadA transports Cd2+ in S. aureus, CopA and CopB transport Cu+ in E. coli, and ZntA
Fig. 1 The schematic model of CPx-type ATPases. The features belonged to P-type
ATPases are shown in ellipses. The phosphate group interacting with aspartate residue of CadA is shown in circle. Other conserved motifs of CPx-type ATPases are also indicated but only in letters. The putative transmembrane segments are drawn in rectangle. cytosolic membrane
N
C
C x x C T G E S x P C T G T K DP
H P G D G x N D x P9
transports Zn2+, Cd2+ and probably Pb2+ in E. coli (Silver et al., 1989; Odermatt et al., 1992; Rensing et al.1997).
1.2.2 The heavy metal transporting CPx-type ATPase
Heavy-metal translocating ATPases including CadA, CopA and CopB contain elements common to all P-type ATPases as well as several unique features only found in the membrane transporters for heavy metals, which are: (1) putative heavy metal-binding sites (CXXC motif) in the polar amino-terminal region; (2) a conserved intramembranous CPC or CPH motif (so called CPx motif); (3) a conserved histidine-proline dipeptide (HP locus) 34 to 43 amino acids carboxyl-terminal to the CPx motif; and (4) a unique number of the membrane-spanning domains (Lutsenko and Kaplan, 1995; Solioz and Vulpe, 1996; Fig. 1). Suggesting that these heavy metal ATPases might form a distinct subgroup of the P-type ATPases. And this subgroup of enzyme family was reclassified as CPx-type ATPases or P1-type ATPases for their sharing the distinctive
features beside those characteristics of P-type ATPases (Lutsenko and Kaplan, 1995; Solioz and Vulpe, 1996).
In addition to CopA, B and CadA, some other heavy-metal translocating ATPases have also been characterized and grouped into CPx-type ATPase enzyme family. For example, the copper transporting MNK protein, in which the mutation of the protein resulted in a human Menkes disease, similar diseases were also found in hamster and mouse (Mercer et al., 1993; Vulpe et al., 1993; Chelly et al., 1993). Another human disease gene, the Wilson’s disease gene, WND, was also a
Bull et al., 1993). Furthermore, a high copper tolerant P-type ATPase, CaCRP1, in Candida albicans (Weissman et al., 2000) was also identified recently and was placed into the CPx-type ATPase family. However, only few of these ATPases has been demonstrated to transport metals, except for CadA transports cadmium, zinc, and lead ions (Tsai et al., 1992; Rensing et al., 1998) and CopB transports copper and silver ions in
Enterococcus hirae (Solioz and Odermatt, 1995), ZntA, a homologue of
CadA in E. coli chromosome, transports cadmium, zinc and lead ions (Beard et al., 1997; Rensing et al., 1997 and 1998; Binet and Poole, 2000), and CopA, involved in the copper homeostasis in E. coli, transports copper ion (Rensing et al., 2000). There were also some
CPx-type ATPases found in other bacteria, but without transport evidence. Using a mixture of oligonucleotides coding for the DKTGT (I/L) T
consensus sequence, specific for the phosphorylation site of this family of ATPases, to screen the genomic Helicobacter pylori library led to
discover a P-type ATPase gene (Melchers et al., 1996). This protein of 686 amino acids carries the consensus sites for phosphorylation and ATP binding and exhibits a 25-30% identity to bacterial Cd2+- and
Cu+-ATPases, together with the postulated N-terminal CXXC ion binding motif and the intramembranous CPC sequences found in CPx-type
ATPases. Others were the cop operons of Helicobacter pylori and
Helicobacter felis, which genes were also cloned by gene library
screening (Bayle et al., 1998) and both operons contain open reading frame for a CPx-type ATPase (CopA) with homology to Cd2+- and Cu+-ATPases including the conserved CXXC and CPC motifs, in which the N-terminatl metal binding property has been reported (Bayle et al., 1998). And more to come to this enzyme family should then be expected
11 in the future.
1.3 The topological study of CPx-type ATPases
Although CPx-type ATPases like CadA has been studied both genetically and biochemically (Nucifora et al., 1989; Tsai et al., 1992; Rensing et al., 1998), more detail investigations in cadmium binding, phosphatase activity and aspartyl kinase activity have yet been defined. For a better understanding of the detail mechanism underlying the enzyme activity of CPx-type ATPase, the knowledge of the structural aspects of these membrane proteins will be essential. Considering of the fact that there will be years before high-resolution crystal structures are determined, the relatively crude two-dimensional models, are still the best strategy to explore the structure-function relationship of a membrane protein like CPx-ATPase. In addition, the two-dimensional "topography", so-called topology map, places major constraints on the possible folding patterns of a membrane protein (Jennings, 1989). The basic idea of integral membrane protein structure in this two-dimensional topography is to predict the transmembrane segments of the polypeptide chain, i.e., the disposition of the chain relative to the lipid bilayer in which it is embedded (Lee and Manoil, 1996).
1.3.1 Inferences from sequence- hydropathy analysis
To set up a topological working model, computer programs were used to analyze the possible transmembrane locations of membrane
was drawn using the hydrophobicity analysis of each individual amino acid within the protein measured by the Goldman-Engelman-Steitz
method (GES-scale) (Engelman et al. 1986) followed by a positive-inside rule (von Heijne 1992).
The GES-scale is calculated from the free energies that require for transferring an amino acid residue in an α helix from the membrane interior to water. It is well known that the hydrophobic interior of a lipid bilayer is an energetically very unfavorable environment for peptide bonds, unless the polar groups are hydrogen bonded (Engelman et al. 1986). For this reason, and because of the tendency of hydrophobic amino acid side chains to partition into lipid bilayer, a
membrane-spanning α helix is an energetically favorable structure for a stretch of hydrophobic amino acid residues (Jennings, 1989).
The hydrophobicity analysis method that originally used is the SOAP program (Kyte and Doolittle, 1982; Klein et al., 1985). It is considered that the hydrocarbon core of membranes is typically 30 Å width, which can be traversed by a α-helix consisting of about 20 residues, and forms a membrane-spanning domain (Kyte and Doolittle 1982). The hydrophobic analysis was usually carried out using the GES-scale
(Engelman et al. 1986) and a window composed of a central 7 or 11 residue was selected. The free energies of the 7 or 11 residues in a window were calculated as a sum or average of all the amino acid residues included. When the position of the window changes from the first residue to the end of the sequence, a plot of the free-energy change versus the residue positions could identify the peaks of high hydrophobic domains. When the peaks of greater than a given criterion level in
13
residues are indicative of potential transmembrane helices.
The positive-inside rule is based on the observation that positive charged amino acids are manifold more abundant in cytoplasmic location, as compared to periplasmic location. This is consistent with positively charged loops serving as cytoplasmic anchors for membrane-spanning regions (von Heijne 1989, 1992).
There were also some other hydropathy systems used to predict transmembrane domains of membrane proteins, such as the TopPred (Von Heijne, 1992; Sipos and Von Heijne, 1993) and TMpred (Hofmann and Stoffel, 1993). All these methods are following the similar principles as described above but more complex. There are examples of hydropathy plot shown in Fig. 2.
1.3.2 Experimental approaches to membrane topology
A variety of biochemical techniques have been employed to elucidate topological structure, including chemical labeling, in situ proteolysis, immunological methods, and molecular genetic approaches (Jennings, 1989). Recently, a method of using the in vitro
transcription/translation technique was also developed to detect the topological structure of certain membrane proteins (Holland and Drickamer, 1986).
The chemical labeling is a well-established and simple method in its principle. It requires a labeling agent that only accesses to one side of the membrane. Labeling agents can be covalently reacting radioactive,
fluorescent, or spin-labeled small molecules, or they can be antibodies or proteolytic enzymes (Jennings, 1989). The most frequently used chemical
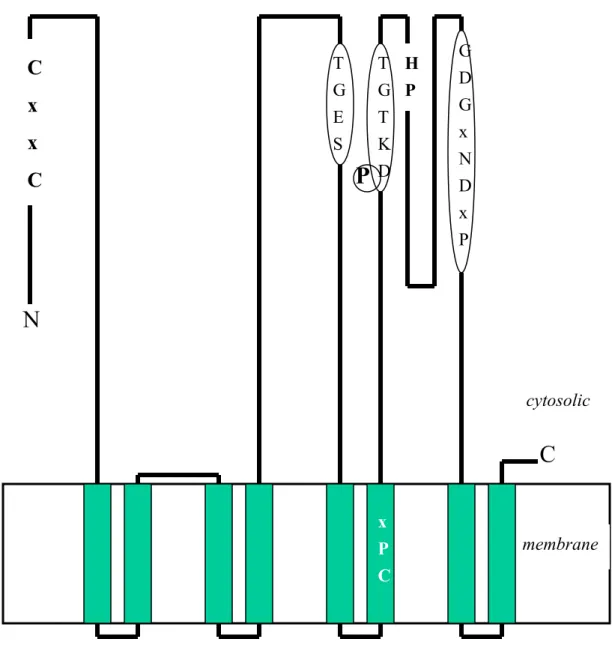
Hydropathy plot of cadA (average of 11 aa) -7 -6 -5 -4 -3 -2 -1 0 1 2 3 1 51 101 151 201 251 301 351 401 451 501 551 601 651 701 Amino acid sequence
Hydr
opathy index
Fig. 2 Hydropathy plots. (a.) Staphylococcal CadA by Kyte and Dollittle method
(SOAP). (b.) Staphylococcal CadA by TMpred program. (c.) H. pylori CadA by TMpred program. The arrows that indicate the peaks of the hydropathy plot are high hydrophobic regions. They usually serve as transmembrane segments in membrane proteins. The black triangles shown in (a.) and (b.) indicate the extra hydrophobic region in staphylococcal CadA but not in H. pylori CadA.
a.
b.
15
is the site-directed chemical labeling of cysteine residues using a
sulfhydryl reagent (Loo and Clarke, 1995). From generating a collection of mutants, each with a single unique cysteine residue, these mutated proteins can be probed with sulfhydryl reagents in oriented membranes to determine the surface accessibility at different residues (Long et al., 1998).
In situ proteolysis method is using the proteolytic enzymes, which
have been used extensively to identify sites that are exposed to the surfaces of membrane proteins. The most common use of this method is to generate and isolate relatively large hydrophobic fragments (Steck et
al., 1976; Jennings et al., 1986). End group determination using the
proteolytic method makes it possible to localize the cleavage site in the primary structure. Proteolytic enzymes are usually used on the outer surface of sealed membranes, or for bilateral cleavage of an unsealed membrane. However, sealing proteolytic enzymes may also work inside RBC ghosts to establish intracellular cleavage sites (Lepke and passow, 1976; Jennings et al., 1986).
Immunological tools have been used increasingly in the study of membrane protein topography recently. In order to be useful for such purposes, the antibody is prepared directly against a specific portion of the primary structure of the protein or an extra epitope inserted to the protein using molecular techniques. The antibodies can be used in topographical work by determining the accessibility of a particular antibody to its target portion of a membrane protein at which side of a sealed membrane. The work of Eckhardt et al. (1999) on mammalian CMP-sialic acid transporter has exemplified this approach in membrane topology studies.
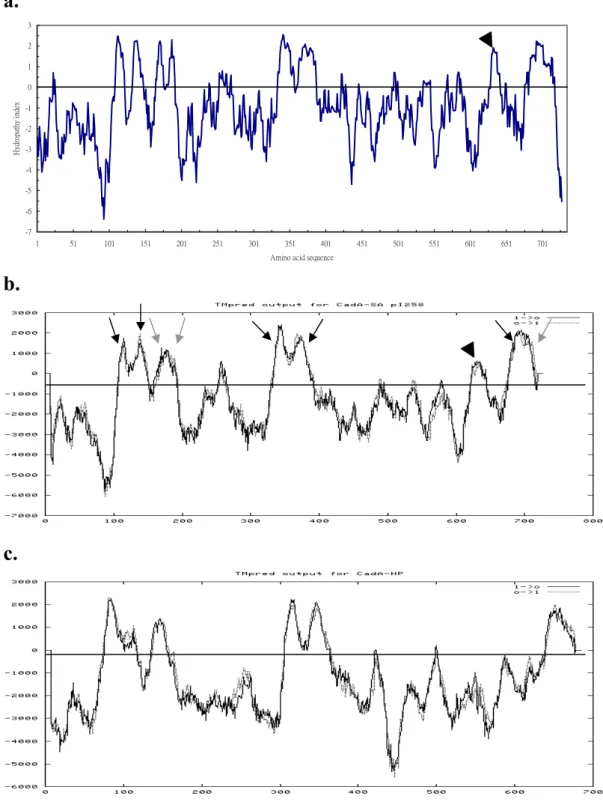
In 1986, Manoil and Beckwith reported a new method for assessing membrane protein topography based on the sidedness of fusion proteins formed from portions of the membrane protein of interest attached to reporter enzyme alkaline phosphatase, which is active only when exported to the periplasm (Hoffman and Wright, 1985). The fusion protein is constructed with the membrane polypeptide upstream from alkaline phosphatase, and the eventual location of the phosphatase will reflect the sidedness of the C-terminus of the membrane protein. Years later, Froshauer et al. (1988) developed a complementary method that employed fusions with β-galactosidase, which is active only in the
cytoplasmic location. On the other hand, fusions to periplasmic C-termini of a membrane polypeptide give low β-galactosidase activity. The
combined uses of periplasmic- and cytoplasmic- fusions provide positive activity signals for the domain determinations on both sides of the
membrane (Manoil et al., 1990; Manoil, 1991, Fig. 3). Hirata et al. (1998) did another example for the combined uses of reporter enzymes in
staphylococcal tetracycline efflux protein, Tet (K), using periplasmic β-lactamase and cytoplasmic chloramphenicol acetyl transferase.
A novel method described for the membrane protein topography was derived from using in vitro transcription/translation of DNA
constructs encoding fusion proteins containing potential transmembrane segments in the presence of microsomes (Holland and Drickamer, 1986). When the fused target peptide is transmembraneous, the
C-terminus-fused signal protein will be transferred into the microsome and glycosylated after in vitro transcription/translation. On the other hand, when the fused peptide is not behind a transmembrane segment, the
17
Fig. 3. Use of fusions to analyze membrane protein topology. Fusion of alkaline
phosphatase to a cytoplasmic membrane protein at a periplasmic site (site 1) yields a hybrid alkaline phosphatase moiety that it is enzymatically active. Fusion of alkaline phosphatase at a cytoplasmic site (site 2) yields an inactive enzyme. β-galactosidase hybrids show reciprocal behavior, with fusions at periplasmic sites yielding proteins with low specific activities when expressed at low levels, which are usually toxic when expressed at high levels. Cytoplasmic sites of β-galactosidase attachment yield high-activity proteins that are relatively nontoxic.
1 2 1 AP+ AP- 2 β-gal- (toxic) β-gal + Periplasmic Cytoplasmic N N N Periplasmic Cytoplasmic
without glycosylation. Using the protein glycosylation or not, the
transmembrane segments could then be defined (Holland and Drickamer, 1986). The topographies of a mammalian P-type ATPase, H+,K+-ATPase (Bamberg and Sachs, 1994), and two CPx-type ATPases from
Helicobacter pylori (Melchers et al., 1996; Bayle et al., 1998) were
established using the approach.
1.3.3 Previous membrane topology studies in CPx-type ATPase related proteins
Two Helicobacter CPx-type ATPases have been determined their topographies using the in vitro transcription/translation method described above (Melchers et al., 1996). Among them, one was suggested as a Helicobacter homologous CadA, since this protein includes a 31% in overall sequence identity among its 686 amino acids to the staphylococcal CadA (Melchers et al., 1996). The amino acid sequence alignment of CadA from S. aureus pI258 and that from H. pylori was shown in Fig. 4. According to Helicobacter topological study, there are 8 transmembrane segments (TM1 to TM8) found in this ATPase (Melchers et al., 1996). Both the N- and C-termini are located in the cytoplasm of the cells. The putative phosphatase domain is between forth and fifth transmembrane segments (TMs) and the phosphorylation site and ATP binding site are both between sixth and seventh TMs in this P-type ATPase (Fig. 1). Other than those, the signatures of CPx-ATPases are also present in this enzyme just as predicted (Solioz and Vulpe, 1996). Later, using alkaline
phosphatase fusion has confirmed the model (Melchers et al., 1999; Fig. 4). The other Helicobacter CPx-type ATPase was suggested as a CopA
19
13
SA pI258 MNVYRVQGFTCANCAGKFEKNVKKIPGVQDAKVNFGASKIDVYGNASVEELEKAGAFENL 72 :. :... : .::.:.:... :. :..::..::. . ..: ..::. :: HP MQEYHIHNLDCPDCASKLERDLNELDYVKKAQINFSTSKL--FLDTS--DFEKVKAF--- 53 1 ion binding site
SA pI258 KVSPEKLANQTIQRVKDDTKAHKEEKTPFYKKHSTLLFATLLIAFG-YLSHFVNGEDNLV 131 .:. ... . : :. : : :. :... .: : .: . HP ---IKQNEPHLSLSFKEATEKPLSF---TPLIITIMVFLGAILILHLNPSPLIE 101 SA pI258 TSMLFVGSIV--IGGYSLFKVGFQNLIRFDF-DMKTLMTVAVIGATIIGKWAEASIVVIL 188 .:.:: ..: ..: ... .:..: . .: : ..:: .:.:.: ..: . :. .... HP KAMFFVLALVYLVSGKDVILGAFRGLRKGQFFDENALMLIATIAAFFVGAYEESVSIMVF 161 SA pI258 FAISEALERFSMDRSRQSIRSLMDIAPKEALVRRNGQEIIIHVDDIAVGDIMIVKPGEKI 248 .. .: :...::..:...:.:.::. : .... . . . .:. :.::..:: :::. HP YSAGEFLQKLAIARSKKSLKALVDVAPNLAYLKKGDELVSVAPEDLRVNDIVVVKVGEKV 221 SA pI258 AMDGIIVNGLSAVNQAAITGESVPVSKAVDDEVFAGTLNEEGLIEVKITKYVEDTTITKI 308 .::..:.: : ... :..:::.::. . ...:..:.:: ....:... :. .:..:.:. HP PVDGVVVKGESLLDERALSGESMPVNVSENSKVLGGSLNLKAVLEIQVEKMYKDSSIAKV 281 Phosphatase domain SA pI258 IHLVEEAQGERAPAQAFVDKFAKYYTPIIMVIAALVAVVPPLFFGGSWDTWVYQGLAVLV 368 . ::..: .:.. .. :. ::..:::: .. :: ..::.:::: ::.: :.:.::..:. HP VDLVQQATNEKSETEKFITKFSRYYTPSVLFIALMIAVLPPLFSMGSFDEWIYRGLVALM 341 SA pI258 VGCPCALVISTPISIVSAIGNAAKKGVLVKGGVYLEKLGAIKTVAFDKTGTLTKGVPVVT 428 :.::::::::.:.. ...: :..::.:.:: :: : :..:::::::::::: :: HP VSCPCALVISVPLGYFGGVGAASRKGILMKGVHVLEVLTQAKSIAFDKTGTLTKGVFKVT 401 Conserved intramembranous motif Phosphorylation site SA pI258 DFEVLNDQVEEKELFSIITALEYRSQHPLASAIMKKAEQDNIPYSNVQ-VEEFTSITGRG 487 :. : . .: :.. . . : ::.: .:.: :. .. . .... ..: : HP DIVPQNGHSKE-EVLHYASCSQLLSTHPIALSIQKACEEMLKDDKHQHDIKNYEEVSGMG 460 Conserved locus SA pI258 IKGIVNGTTYYIGSPKLFKELNVSDFSLGFENNVKILQNQGKTAMIIGTEKTILGVIAVA 547 .:. . :. :.. ... : . ::. : . .. ..: .: :... HP VKAQCHTDLIIAGNEKMLDQFHIAH-SPSKENG---TIVHVAFNQTYVGYIVIS 510 SA pI258 DEVRETSKNVIQKLHQLGIKQTIMLTGDNQGTANAIGTHVGVSDIQSELMPQDKLDYIKK 607 ::... . . .. :. ::.. .:.:: ...:. .: . .. :.:..: . .: HP DEIKDDAIECLRDLKVQGIENFCILSGDRKSATESIAQTLGC-EYHASLLPEEKTSVFKT 569 SA pI258 MQSEYDNVAM-IGDGVNDAPALAASTVGIAMGGAGTDTAIETADIALMGDDLSKLPFAVR 666 .. .: :. .:::.::::.::.. :::.:: :.. . ..:::.. .:.:..: .. HP FKERYKAPAIFVGDGINDAPTLASADVGIGMG-KGSELSKQSADIVITNDSLNSLVKVLA 628 ATP binding domain
SA pI258 LSRKTLNIIKANITFAIGIKIIALLLVIPGWLTLWIAILSDMGATILVALNSLRLMRVKDK 727 ...:: .:: :: ::.::: . ..: . : .:: :...:.:.:.:. ::.: ::.
HP IAKKTKSIIWQNILFALGIKAVFIVLGLMGVASLWEAVFGDVGVTLLALANSMRAMRA 686
Fig. 4. Comparison of S. aureus CadA and the homologous H. polori ATPase. The amino
acid sequences of CadA homologous pumps are 727 (S. aureus pI258) and 686 (H. pylori) amino acids in length. The overall sequence identity level between the two pumps is about 31%. The occurrence of transmembrane segments as experimentally determined in the H.
pylori pump by in vitro method (Melchers, 1996) are highlighted; by in vivo alkaline
phosphatase fusions (Melchers, 1999) are indecated by triangles. That corresponding to the conserved motifs characteristic for CPx-type ATPases are boxed.
homology, based on the reasons of the high sequence similarity to
Enterococcus hirae CopA and the special Cu2+ binding affinity to the N-terminal peptide of this ATPase (Bayle et al., 1998). As determined experimentally by in vitro transcription/translation method, H. pylori CopA also contains four pairs of TMs, and shares the same patterns of motifs with the predicted Helicobacter CadA.
Alignment of all the P-type ATPases suggests that the Helicobacter pumps and other homologous heavy metal ATPases have an additional pair of membrane sequences preceding the first pair of membrane segments that are present in the non-heavy metal ATPases. The
Helicobacter pumps and other similar P-type ATPases, on the other hand, do not have sequences following the TM8 (Melchers et al., 1996). The heavy metal transporting CPx-type ATPases set themselves apart from the rest of the P-type ATPases not only at the sequence aspect, but also at the membrane topology concerns (Solioz and Vulpe, 1996).
1.4 Motivations for CadA topology
Although the topological study of CPx-type ATPases has been done in Helicobacter models, some uncertainties were remained. Firstly, the experimental data presented to this topology model for this transition metal ATPase was from in vitro study and only 9 in vivo phoA fusion data in a subsequent study (Melchers et al., 1996; Melchers et al., 1999; Fig. 4). However, there are still 3 large cytoplasmic domains of this protein has not been explored, which are the N-terminal first 70 residues, and the regions ranging from amino acid positions 177 to 309 and 371 to 493. Additionally, the third and forth TMs (TM3 and TM4) suggested in these
21
studies were originally predicted to be as low hydrophobic domains, according to hydropathy programs (Fig. 2c). It seems that they were not able to traverse the lipid bilayer. In the in vitro study, the suggested TM4 of the CadA homologous ATPase was not present as completed
transmembrane domain (Melchers et al., 1996). The in vivo alkaline phosphatase fusions were too few to cover the full length of the protein including this region (TM3 to TM4) (Melchers et al., 1999). Therefore, this information would not be possible to serve as the perfect topological model for the CPx-type ATPases.
In addition to the reasons we have argued above, some differences exist between these two ATPases. From the sequence comparison data shown in Fig. 4, staphylococcal CadA has a longer N-terminal portion of amino acid sequences and more charged C-terminal portion of amino acid residues than those of the Helicobacter one. Moreover, some CPx-type ATPases including staphylococcal CadA have another putative
transmembrane domain, just behind the GDGXNDXP conserved motif, which were not predicted as a transmembrane domain in the ATPases of
Helicobacter spp (Fig. 2). The similar domain was also shown in other
CPx-type ATPases like CadA ATPases from S. aureus Tn554, Bacillus
firmus, Listeria monocytogenes as well as PacS and CtaA from
Synechococcus spp (Chikramane and Dubin, unpublished; Ivey et al.,
1992; Lebrun et al., 1994; Kanamaru et al., 1993; Phung et al., 1994). Therefore, another membrane topology data would be necessary to clarify these suspicions.
Since the topography acquired from Helicobacter studies were not completed to truly represent the CPx-type ATPases, therefore, it will be necessary to re-examine the protein structure of the family. In this thesis study, we decide to use CadA as a model to further disclose the structural aspect of the CPx-type ATPase. There are many experimental techniques would be able to study the topological structure of membrane proteins, including side chain covalent modification, proteolysis, and epitope recognition (Jennings, 1989). However, these techniques are often unable to provide a full topological description of a membrane protein because of their reliance on the natural occurrence of residues that are susceptible to proteolytic cleavage, reactive side chains or accessible to antibody binding (Lee and Manoil, 1996). Another method, the in vitro
transcription/translation technique, seems amazing (Holland and
Drickamer, 1986), but difficult to reflect the intact protein folding in cell membrane because of using only those putative transmembrane fragments for detection. These limitations could be circumvented by the supplement with data from some other molecular biological methods, especially those of gene fusion strategies. The best-established gene fusion strategy for the study of this kind is the phoA- and lacZ-fusion combination method
(Manoil et al., 1990; Manoil, 1991; Fig. 3). This method has been used in many membrane topological studies, for example, the E. coli ArsB done by Wu et al. (1992), E. coli K-12 Mtr permease done by Sarsero and Pittard (1995), B. subtilis BofA done by Varcamonti et al. (1997),
Lactococcal LcnC done by Franke et al. (1999), Helicobacter NixA
done by Fulkerson and Mobley (2000). In this study, we take the similar strategy of using the phoA- and lacZ-fusion to determine the membrane topography of CadA for the first time.
23
1.4.2 Properties of reporter enzymes
Alkaline phosphatase is a nonspecific phosphomonoesterase, which has a total of 471 amino acids including a signal sequence of 21 amino acids (Chang, 1986). It is ordinarily found in the periplasm of E. coli under the condition of phosphate starvation (Wanner 1996). If retained in the cytoplasm, alkaline phosphatase is enzymatically inactive (Michaelis 1983, Hoffman 1985), due to the inability of forming disulfide bonds (Derman and Beckwith, 1991). This property of alkaline phosphatase forms the basis for a convenient in vivo strategy for monitoring protein translocation across the cytoplasmic membrane (Manoil et al. 1990).
On the other hand, β-galactosidase is a tetramer, and each identical monomer is 116,353 Dalton in molecular weigth and 1,023 amino acid residues in length (Fowler and Zabin, 1978). It is active normally in the cytoplasm (Manoil et al., 1988; Slauch and Silhavy, 1991), but inactive or even toxic when secreted to the periplasm of E. coli (Snyder and Silhavy, 1995).
Using these two differently orientated reporter genes to fuse with membrane protein at different locations, completed membrane protein topography will then be possible (Froshauer et al., 1988).
1.5Rationale and experimental approach
The main purpose of this thesis study is to characterize the topography of CadA protein. To determine how CadA spans on the plasma membrane of bacteria cells; how many TMs are involved in this
protein; where the amino and carboxyl termini are located; and which side the functional groups or the conserved motifs are placed.
Furthermore, it is expected to provide a better topological model for CPx-type ATPases from this study.
Special arm:
1. To determine the computer predicted CadA hydrophathy.
2. Using alkaline phosphatase and β-galactosidase as the reporter genes to determine the presence of transmembrane segments in CadA.
3. Prepare a detail CadA topological model using the enzymatic data provide from this study.
4. Disclose the similarities and differences between the staphylococcal CadA and its homologous Helicobacterial CadA in this study.
25
Chapter 2. Materials and Methods
2.1 Bacterial strains, plasmids, and culture conditions
E. coli strains and plasmids used in this study are described in Table
I. E. coli strain JM109 was used as host for gene cloning purpose. The E.
coli strain LMG194 was used for phoA fusion protein expression, and
strain MC1000 was used for lacZ fusion protein expression. In all experiments indicated in this study, bacterial cells were grown in LB medium (1% tryptone, 1% NaCl, and 0.5% yeast extract in water) at 37ºC with 200 rpm shaking and supplement with 100μg/ml of ampicillin. A 400μg of either 5-bromo-4-chloro-3-indolyl phosphate (BCIP) or 5-bromo-4-chloro-3-indolyl β-D-galactopyranoside (X-GAL) were spread onto LB agar plates (1.5% agar in LB medium) for screening alkaline phosphatase or β-galactosidase containing bacterial clones.
2.2 Molecular biology techniques
Gene cloning was performed using the standard procedures and
described as follow. Polymerase Chain Reactions (PCR) were preformed using Takara Ex Taq (Takara) enzyme in a PTC-200 thermocycler (MJ Research). Plasmids and fragment DNAs, including PCR products and digested DNA, were purified using NucleoSpin Plus Plasmid Miniprep Kit and NucleoSpin Extraction Kit (Clontech), respectively. Restriction enzymes were purchased from New England Biolabs and DNA ligation reactions were performed using a T4 DNA ligase (Promega).
Table I Strains and plasmids
Strain/plasmid Genotype/description Refercnce
E. coli strains
JM109
LMG194 MC1000
RW3110
F' traD36 lacIq ∆(lacZ)M15 proA+B+ /
e14-(McrA- )∆(lac- proAB) thi gyrA96 (NaIr) endA1
hsdR17(rK- mK+) recA1 relA1 supE44
F- ∆(lacIPOZY)X74 galE galK thi rpsL ∆phoA ra714
F- araD139 ∆(araABC-leu)7679 galU galK ∆(lac)X74
rpsL thi
F- mcrA mcrB IN(rrnD-rrnE)1 lambda- ZntA::km
Promega Invitrogen Ellis et al. 1995 Rensing et al. 1997 Plasmids pKJ3 pFU3K pSE380 pMM1 pMLB1069 pKJ100 pXxP series pXxL series
2.6-kilobase pair XbaI fragmeng containing the 3' end of cadC and the complete cadA gene from S. aureus in pET11a
entire cadA gene, 2184 base pair NcoI-EcoRI fragment without internal NcoI and EcoRI, ligated into
pBADMycHisA
an expression vector offering trc promoter, lacO operator, lacIq repressor and AmpR selection marker a transposon delivery plasmid carrying a
mini-transposon TnTAP and Tn5 transposase pBR322 ∆(tetr) lac’ZY’
NcoI- XbaI fragment of entire cadA gene in vector
plasmid pSE380
Fusions of phoA to various PCR fragments of cadA gene
Fusions of lacZ to various fragments of cadA
K. J. Tsai S. L. Fu Invitrogen Guan et al. 1999 Ellis et al. 1995 This study This study This study
27 (Bio-Rad).
2.3 Construction of inducible cadA expression system
In order to obtain a well-controlled cadA expression system, the
plasmid pSE380 (Invitrogen) was chosen as the vector for cloning
purpose. The pSE380 was digested with XbaI and HindIII, and then filled with a 368-bp XbaI-HindIII fragment from pET11a to yield pSE380a plasmid, in which the SpeI site was removed. A 2.2-kb NcoI-EcoRI cadA gene from pFU3K was cut out and ligated into NcoI-EcoRI digested pSE380a to make pSE-FU3K plasmid. Then replaced the 1.7kb
SpeI-XbaI fragment of pSE-FU3K with a SpeI-XbaI fragment from pKJ3
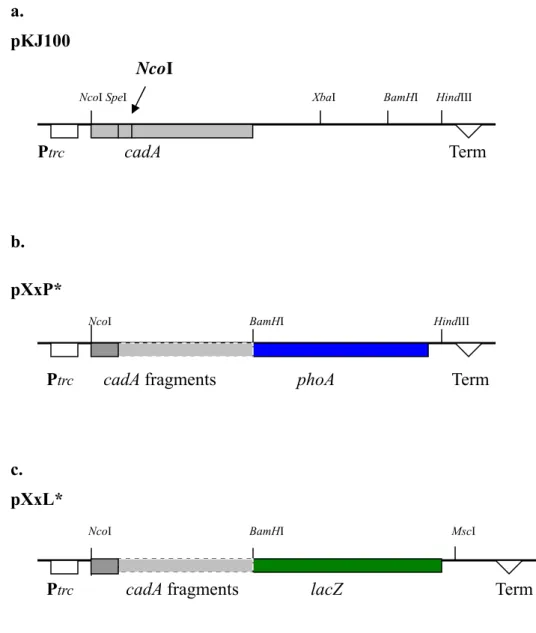
to generate plasmid pKJ100 (Fig. 5a). In this construction, an
approximately entire cadA gene is controlled by the trc promoter, which is a hybrid promoter from trp and lac operon.
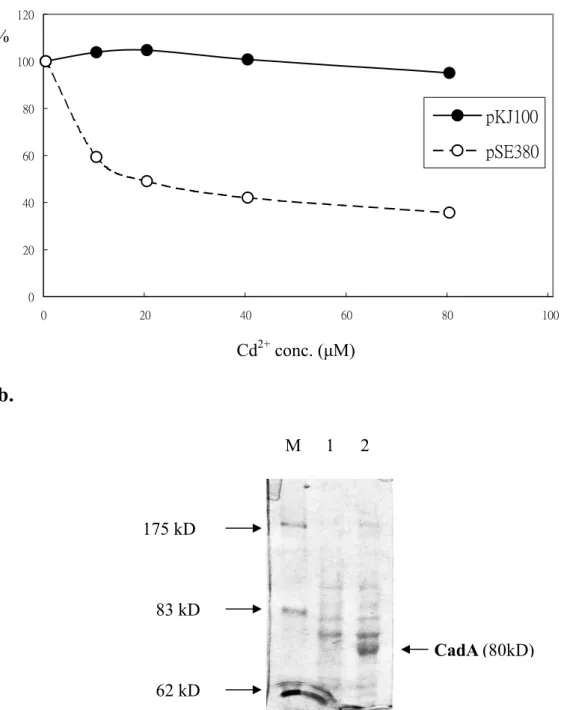
2.4 Cadmium resistance assays
Cd2+ resistance assays were performed using the method previously described (Nucifora et al. 1989). RW3110 cells harboring pKJ100 or pSE380 were cultured in LB medium in the presence of 100 μg/ml of ampicillin at 37ºC overnight. Aliquots of cultures were diluted 1: 50 with fresh LB containing 0, 10, 20, 40, and 80 μM CdCl2, respectively and
grew for another 6 hours. The cell survivals were measured spectrometrically at a wavelength of 600 nm.
a.
pKJ100
NcoI
NcoI SpeI XbaI BamHI HindIII
Ptrc cadA Term
b.
pXxP* NcoI BamHI HindIII
Ptrc cadA fragments phoA Term
c. pXxL*
NcoI BamHI MscI
Ptrc cadA fragments lacZ Term
Fig. 5. Constructed plasmids. (a.) The plasmid pKJ100 contains the full-length cadA
gene. (b.) The plasmids contain various cadA-phoA fusions. (c.) The various plasmids contain the cadA-lacZ fusions. The vector plasmid used for these constructions was pSE380. The regulation pattern of this vector is the trc promoter, lac operator, and it’s own terminator. Restriction sites used in construction were also shown. An internal
NcoI site in the cadA gene was marked. * The capital "X" represents the residue name
on CadA protein just before the fusion site. The lowercase "x" represents this residue sequence number.
29
2.5 Construction of cadA-phoA and cadA-lacZ fusions
Either phoA or lacZ fusion with the truncated cadA were prepared by
ligating the PCR fragments of cadA to the 5'-end of these reporter genes, as described below
2.5.1 CadA-phoA fusion
The cadA-phoA gene fusions were constructed in vitro and cloned in
E. coli strain JM109. The signal sequenceless phoA gene in the 1.9-kb BamHI-HindIII fragment from pMM1 (Ehrmann et al., 1997; Guan et al.
1999) was cut out and ligated to pSE380 digesting with the same
enzymes, producing the pSE-phoA clones. PCR fragments, summing up 22 different lengths of cadA gene, were produced using pKJ3 as template. And used the same positive-strained primer and different
negative-strained primers (Tab. IIa). The first 6 PCR products that are without internal NcoI site were digested with NcoI and BamHI and cloned into digested pSE-phoA to generate clones pL109P, pL130P, pL155P, pE181P, pR204P and pV326P. The rest of the 16 fragments were digested with SpeI and BamHI to prevent the NcoI sites in these PCR fragments and placed onto the pV326P plasmid digested with the same enzymes. Totally, 22 cadA-phoA fusions were achieved, and named them as pXxP according to the residue name (X) and number (x) before the fusion junction sites of cadA-phoA fusions (Fig. 5b).
Table II Primers. (a.) Primers used in PCR that for cadA-phoA and cadA-lacZ
constructions. The numbers in the primer names correspond to the DNA sequence of
cadA. The added restriction sites are underlined. Altered nucleotides from cadA gene
are written in lowercase letters. (b.) Primers used for DNA sequencing.
a. NcoI(+) BamHI327(-) BamHI390(-) BamHI465(-) BamHI543(-) BamHI612(-) BamHI690(-) BamHI765(-) BamHI849(-) BamHI978(-) BamHI1068(-) BamHI1164(-) BamHI1242(-) BamHI1335(-) BamHI1431(-) BamHI1527(-) BamHI1626(-) BamHI1731(-) BamHI1833(-) BamHI1938(-) BamHI2043(-) BamHI2109(-) BamHI2178(-) 5' GTGAAGGTCcATGgCTGAACAAAAG 3' 5' GTGTGGgAtcCAGCAATGTACTATG 3' 5'CATGGAAGgatCCAGGTTATCTTCTCC 3' 5' AATCAAAGgGatcCAAATTTTGAAA 3' 5' ACAACAATAGgatCCTCTGCCC 3' 5' GAACGTATGGgaTccCTTGATCTGTCC 3' 5' CCCACAGCGATAggaTCCACATGG 3' 5' GCCGACAAGggATcCACAATGATTCC 3' 5' TCGTTAAGCGgAtCcGCAAATACTTC 3' 5' CGCAAATggATCcACGAATGCTTGGG 3' 5' AAACCCATGgATCCCAACTG 3' 5' TTTTTTCGCTGgATccCCAATTGCCG 3' 5' TTCCTGTTggATCcAATGCGACTGTC 3' 5' CTAAAGCTGgAtccATAGAGAATAGCTC 3' 5' CGAAGTGAATgaaTCCACTTGTAC 3' 5' GGCTAAAATCGGgAtCcTTTAATTCC 3' 5' CATCTGCAACGGgAtccACGCCGAGAATTG 3' 5' GCATTTGCAGgAtCcTGATTATCACC 3' 5' GCTACATTAggATcCTCCGATTGC 3' 5' ATCAGCTGgaTCcATTGCAGTATCCG 3' 5' TTTAATTCCGggAtCcAAAGTGATG 3' 5' ATATCGGAAgGAtcCGCTATCCAAAG 3' 5' TCTACCTATggATCCTTCACTC 3' b. SE204F AP113R LAC1362R 5' CAATTAATCATCCGGCTCG 3' 5' GCAGTAATATCGCCCTGAGCAGC 3' 5' GGGGATGTGCTGCAAGGCG 3'
31
2.5.2 CadA-lacZ fusion
The cadA-lacZ gene fusions were obtained following the similar procedure as that of cadA-phoA fusions. A 3.3-kb BamHI-MscI lacZ fragment from plasmid pMLB1069 (Ellis et al. 1995) was prepared to replace the BamHI-MscI region of the pSE380 plasmid to generate the pSE-lacZ plasmid. Similarly, the same 22 PCR fragments as mentioned above were cloned separately into this vector using either NcoI-BamHI or
SpeI-BamHI digestion as described and named these clones as pXxL (Fig
5c). The plasmids are called pXxL followed the residue name and number before the fusion junction sites.
2.6 DNA Sequencing
All selected clones were subjected to DNA sequence determination using an ABI PRISMTM Dye Terminator Cycle Sequencing System in core facility laboratory in this school. The sequencing primers used in these sequencing reactions were listed in the Table IIb.
2.7 Expression of the hybrid proteins
Plasmids that contained cadA-reporter gene fusions were
transformed into either LMG194 for phoA fusions, or MC1000 for lacZ fusions, using electroporation for hybrid protein expressions.
Transformed cells were cultured in 5 ml of LB medium in the presence of 100 μg of ampicillin at 37ºC overnight with 200 rpm shaking. Next
continuously grow until an optical density of A600 nm between 0.5 and 0.6.
A 0.1 mM Isopropyl β-D-Thiogalactopyranoside (IPTG) was then added to the culture to induce the protein expression at 30ºC for two hours. Finally, the cells were harvested and proteins were prepared for reporter enzyme activity assays and Western blotting analysis. In each culture, a portion of culture was withdrawn for mini-prep DNA preparation to confirm the presence of each hybrid gene using restriction fragment gel electrophoresis.
2.8 Enzyme activity assays
Alkaline phosphatase and β-galactosidase activity assays were
performed using the methods of Manoil's (1991) and Miller's (1972), respectively. Some modification were made as described below. The data was collected and calculated from three to five independent experiments.
2.8.1 Alkaline phosphatase
The CadA-alkaline phosphatase fusion proteins were expressed in LMG194 cells as described above. Cells were harvested by centrifugation and washed once with Tris-HCl (pH8.0) buffer. Pellets were resuspended in 1 ml of the same buffer in the presence/absence of one drop of 0.01% SDS and chloroform. A 0.1 ml of the samples was mixed with 0.9 ml of AP buffer containing 100 mM NaCl, 5 mM MgCl2, in 100 mM Tris-HCl
(pH 9.5) and read in a spectrophotometer. The values of A600 nm were
recorded as the reference reading for the assay. Another 0.1 ml of the samples were mixed with 0.85 ml of AP buffer and 0.05 ml of 0.4%