CHAPTER 2
DNA: The Genetic
Material
台大農藝系 遺傳學 601 20000 Chapter 2 slide 2
The Search for the Genetic Material
• 1. Some substance must be responsible for
passage of traits from parents to offspring. For a substance to do this it must be:
• a. Stable enough to store information for long periods.
• b. Able to replicate accurately.
The Search for the Genetic Material
• 2. In the early 1900s, chromosomes were shown
to be the carriers of hereditary information. In eukaryotes they are composed of both DNA and protein, and most scientists initially believed that protein must be the genetic material.
台大農藝系 遺傳學 601 20000 Chapter 2 slide 4
Chromosome theory of inheritance
• The transmission and behavior of the abstract
factors of Mendel and the physical behavior of
chromosomes in mitosis, meiosis, and fertilization led Sutton, Roux and Boveri proposed the
Genetic Materials
• Chromosome consists of protein and nucleic acid • Candidate: Protein v.s. nucleic acid
• Protein: 20 kinds of amino acid
• Nucleic acid: 4 kinds of nucleotides
• Complexity of life very complicated protein
or nucleic acid to account for the level of complexity?
台大農藝系 遺傳學 601 20000 Chapter 2 slide 6
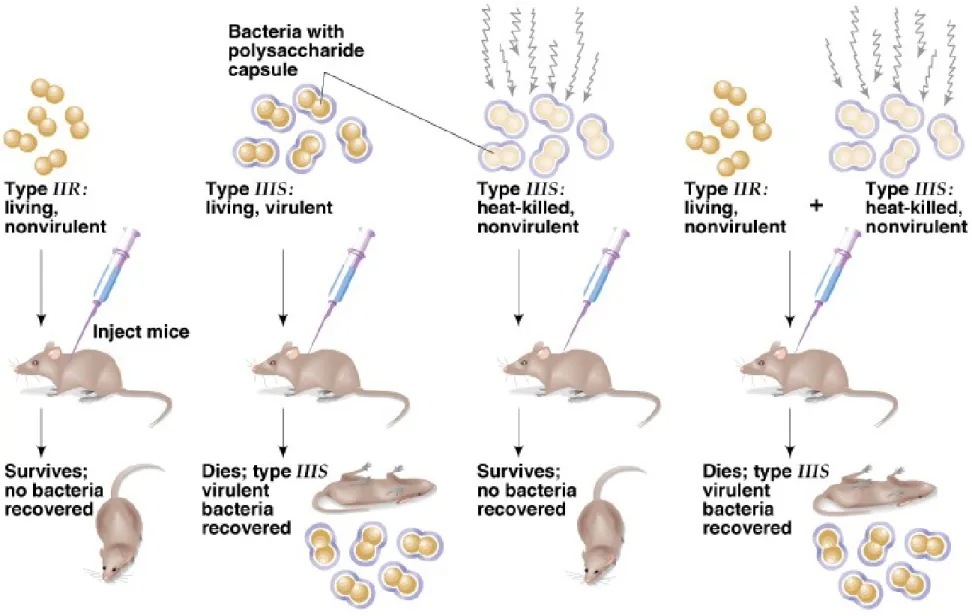
Griffith’s Transformation Experiment
• 1. Frederick Griffith’s 1928 experiment with
Streptococcus pneumoniae bacteria in mice
showed that something passed from dead bacteria into nearby living ones, allowing them to change their cell surface.
• 2. He called this agent the transforming
principle, but did not know what it was or how it worked.
台大農藝系 遺傳學 601 20000 Chapter 2 slide 8
Avery’s Transformation Experiment
• Animation: DNA as Genetic Material: The Avery Experiment
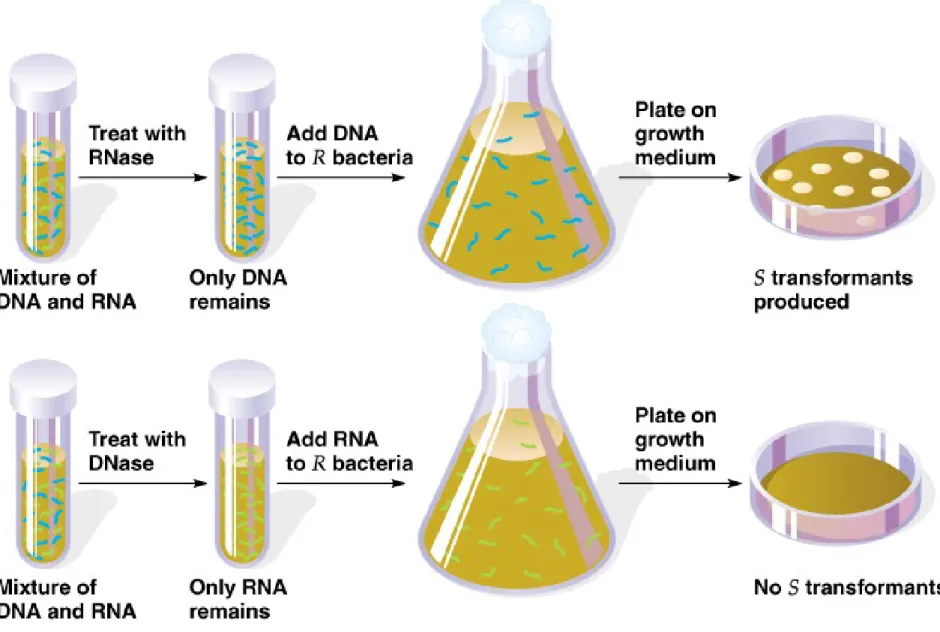
• 1. In 1944, Avery, MacLeod and McCarty published results of a study
that identified the transforming principle from S. pneumoniae. Their approach was to break open dead cells, chemically separate the
components (e.g., protein, nucleic acids) and determine which was capable of transforming live S. pneumoniae cells.
• 2. Only the nucleic acid fraction was capable of transforming the bacteria.
• 3. Critics noted that the nucleic acid fraction was contaminated with
proteins. The researchers treated this fraction with either RNase or protease and still found transforming activity, but when it was treated with DNase, no transformation occurred, indicating that the
Fig. 2.3 Experiment that showed that DNA, not RNA, was the transforming principle
台大農藝系 遺傳學 601 20000 Chapter 2 slide 10
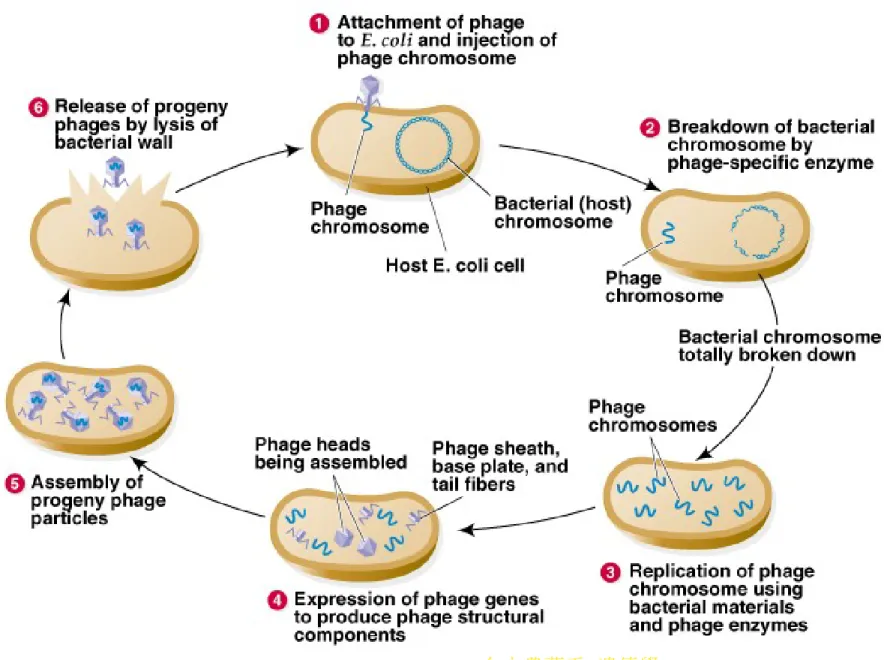
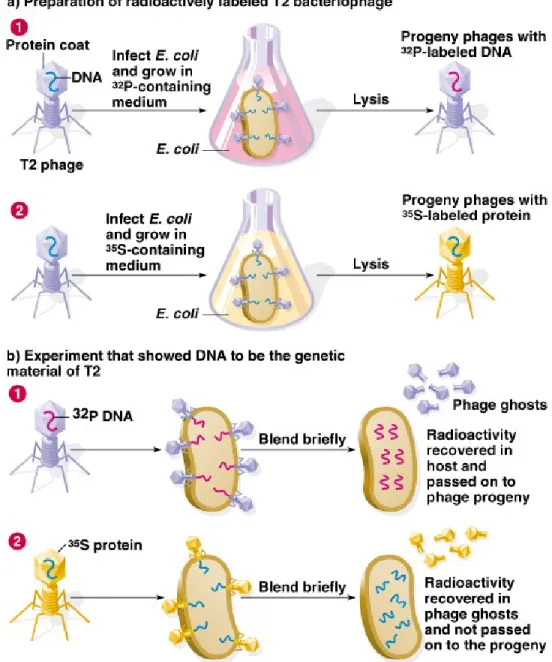
The Hershey-Chase Bacteriophage Experiment
• Animation: DNA as Genetic Material: The Hershey-Chase Experiment • 1. More evidence for DNA as the genetic material came in 1953 with
Alfred Hershey and Martha Chase’s work on E. coli infected with bacteriophage T2.
• 2. In one part of the experiment, T2 proteins were labeled with 35S, and
in the other part, T2 DNA was labeled with 32P. Then each group of
labeled viruses was mixed separately with the E. coli host. After a short time, phage attachment was disrupted with a kitchen blender, and the location of the label determined.
• 3. The 35S-labeled protein was found outside the infected cells, while
the 32P-labeled DNA was inside the E. coli, indicating that DNA carried
the information needed for viral infection. This provided additional support for the idea that genetic inheritance occurs via DNA.
台大農藝系 遺傳學 601 20000 Chapter 2 slide 12
Peter J. Russell, iGenetics: Copyright © Pearson Education, Inc., publishing as Benjamin Cummings.
台大農藝系 遺傳學 601 20000 Chapter 2 slide 14
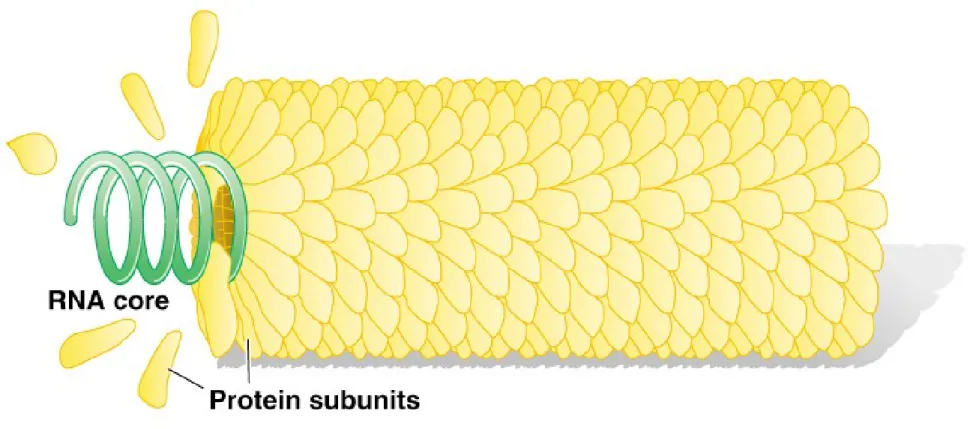
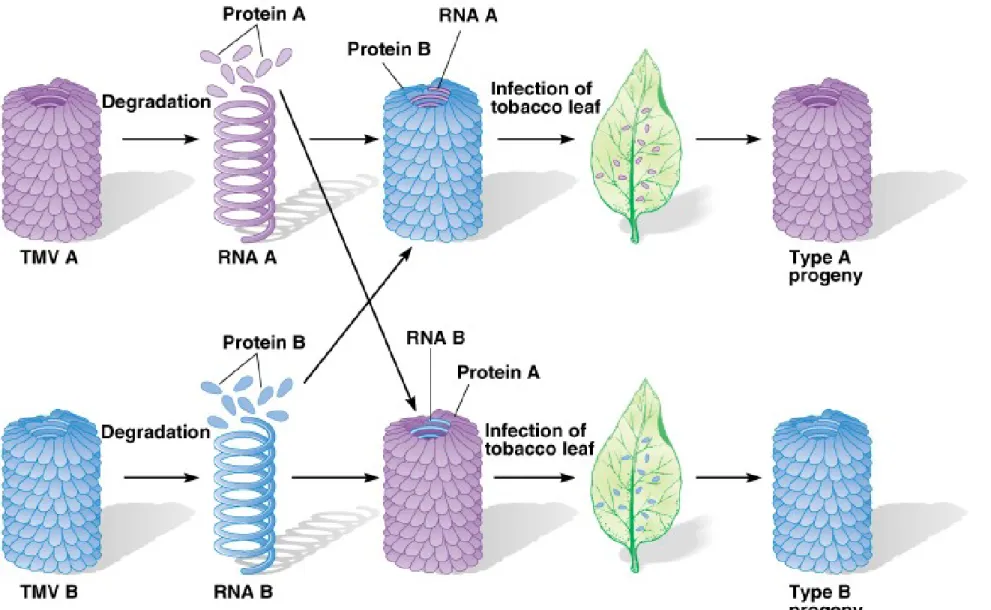
The Discovery of RNA as Viral Genetic Material
• TMV ( tobacco mosaic virus)
• 1956, A. Gierer and G. Schramm
• Infected tobacco plant with purified RNA typical virus-infected lesion
• RNA treated with RNAase then injected into tobacco not lesion
• 1957 Heinz Fraenkel-Conrat and B. Singer reconstitue the RNA of one type with the protein of the other type and
vice versa and injected to two tobacco plants the
progeny viruses isolated from the resulting lesion were the type specified by the RNA, not by the protein.
台大農藝系 遺傳學 601 20000 Chapter 2 slide 16
Peter J. Russell, iGenetics: Copyright © Pearson Education, Inc., publishing as Benjamin Cummings.
Fig. 2.8 Demonstration that RNA is the genetic material in tobacco mosaic virus (TMV)
The Composition and Structure of DNA and
RNA
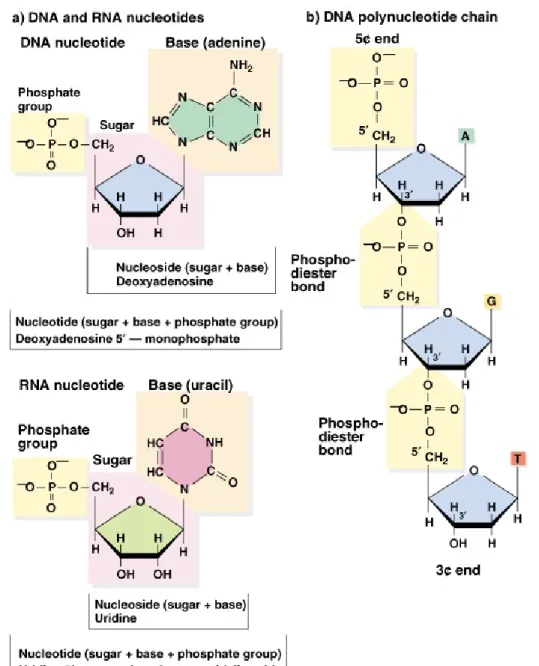
• 1. DNA and RNA are polymers composed of monomers
called nucleotides.
• 2. Each nucleotide has three parts:
• a. A pentose (5-carbon) sugar.
• b. A nitrogenous base.
• c. A phosphate group.
• 3. The pentose sugar in RNA is ribose, and in DNA it’s
deoxyribose. The only difference is at the 29 position, where RNA has a hydroxyl (OH) group, while DNA has only a hydrogen.
台大農藝系 遺傳學 601 20000 Chapter 2 slide 18
Peter J. Russell, iGenetics: Copyright © Pearson Education, Inc., publishing as Benjamin Cummings.
The Composition and Structure of DNA and
RNA
• 4. There are two classes of nitrogenous bases:
• a. Purines (double-ring, nine-membered structures) include adenine (A) and guanine (G).
• b. Pyrimidines (one-ring, six-membered structures)
include cytosine (C), thymine (T) in DNA and uracil (U) in RNA.
台大農藝系 遺傳學 601 20000 Chapter 2 slide 20
Peter J. Russell, iGenetics: Copyright © Pearson Education, Inc., publishing as Benjamin Cummings.
The Composition and Structure of DNA and
RNA
• 5. The structure of nucleotides has these features:
• a. The base is always attached by a covalent bond between the 1′
carbon of the pentose sugar and a nitrogen in the base (specifically, the nine nitrogen in purines and the one nitrogen in pyrimidines).
• b. The sugar-base combination is a nucleoside. When a phosphate is added (always to the 5′ carbon of the pentose sugar), it becomes a nucleoside phosphate, or simply nucleotide.
• c. Nucleotide examples are shown in Figure 2.11, and naming conventions are given in Table 2.1.
• 6. Polynucleotides of both DNA and RNA are formed by stable
covalent bonds (phosphodiester linkages) between the phosphate group on the 5′ carbon of one nucleotide, and the 3′ hydroxyl on another
nucleotide. This creates the “backbone” of a nucleic acid molecule.
• 7. The asymmetry of phosphodiester bonds creates 3′-5′ polarity within the nucleic acid chain.
台大農藝系 遺傳學 601 20000 Chapter 2 slide 22
Peter J. Russell, iGenetics: Copyright © Pearson Education, Inc., publishing as Benjamin Cummings.
The Discovery of the DNA Double Helix
• 1. James Watson and Francis Crick published the famous double-helix structure in 1953. When they began their work, it was known that DNA is composed of
nucleotides, but how the nucleotides are assembled into nucleic acid was unknown. Two additional sources of data assisted Watson and Crick with their model:
• a. Erwin Chargaff’s ratios obtained for DNA derived from a
variety of sources showed that the amount of purine always
equals the amount of pyrimidine, and further, that the amount of G equals C, and the amount of A equals T.
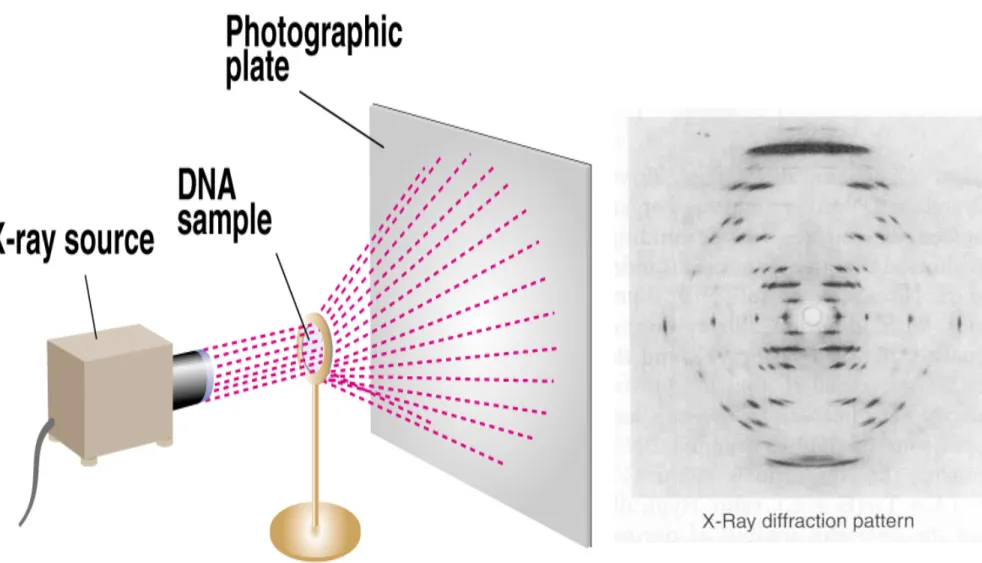
• b. Rosalind Franklin’s X ray diffraction images of DNA showed a helical structure with regularities at 0.34 nm and 3.4 nm along the axis of the molecule (Figure 8.9).
台大農藝系 遺傳學 601 20000 Chapter 2 slide 26
Peter J. Russell, iGenetics: Copyright © Pearson Education, Inc., publishing as Benjamin Cummings.
台大農藝系 遺傳學 601 20000 Chapter 2 slide 28
The Discovery of the DNA Double Helix
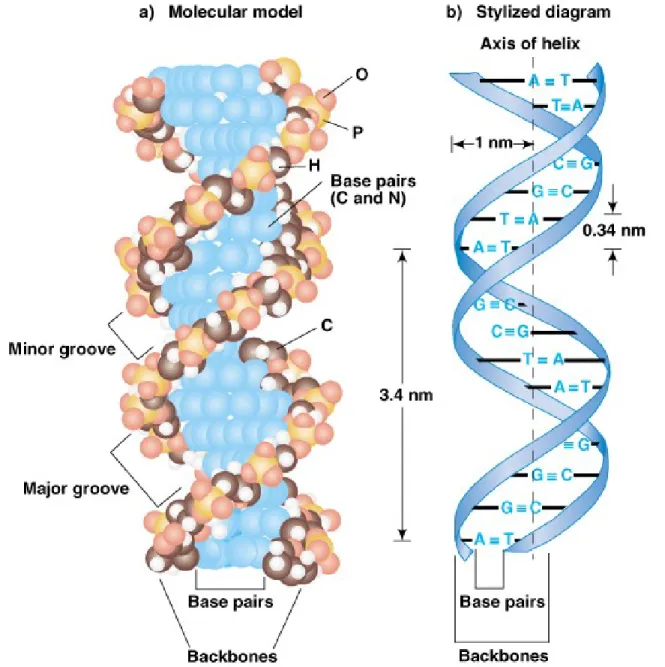
• 2. Watson and Crick’s
three-dimensional model (Figure 2.14) has these main features:
• a. It is two polynucleotide chains wound around each other in a right-handed helix.
• b. The two chains are antiparallel.
• c. The sugar-phosphate backbones are on the outside of the helix, and the bases are on the inside,
stacked perpendicularly to the long axis like the steps of a spiral staircase.
• d. The bases of the two strands are held together by hydrogen bonds between complementary bases (two for A-T pairs and three for G-C pairs). Individual H-bonds are relatively weak and so the strands can be separated (by heating, for example). Complementary base pairing means that the
sequence of one strand dictates the sequence of the other
台大農藝系 遺傳學 601 20000 Chapter 2 slide 30
• e. The base pairs are 0.34 nm apart, and one full turn of the DNA helix takes 3.4 nm, so there are 10 bp in a complete turn. The
diameter of a dsDNA helix is 2 nm.
• f. Because of the way the bases H-bond with each other, the opposite sugar-phosphate backbones are not equally spaced, resulting in a major and minor groove. This feature of DNA structure is important for protein binding.
• 3. The 1962 Nobel Prize in Physiology or Medicine was
awarded to Francis Crick, James Watson and Maurice Wilkins (the head of the lab in which Franklin worked). Franklin had already died, and so was not eligible.
Different DNA Structures
• X ray diffraction studies show that DNA can exist in different forms (Figure 2.16).
• a. A-DNA is the dehydrated form, and so it is not usually found in cells. It is a right-handed helix with 10.9 bp/turn, with the bases inclined 13° from the helix axis. A-DNA has a deep and narrow major groove, and a wide and shallow minor groove.
• b. B-DNA is the hydrated form of DNA, the kind normally found in cells. It is also a right-handed helix, with only 10.0 bp/turn, and the bases inclined only 2° from the helix axis. B-DNA has a wide major groove and a narrow minor groove, and its major and minor grooves are of about the same depth.
• c. Z-DNA is a left-handed helix with a zigzag sugar-phosphate backbone that gives it its name. It has 12.0 bp/turn, with the
bases inclined 8.8° from the helix axis. Z-DNA has a deep minor groove, and a very shallow major groove. Its existence in living
DNA in the Cell
• All known cellular DNA is in the B form. • A-DNA would not be expected because it is
dehydrated and cells are aqueous.
• Z-DNA has never been found in living cells,
although many organisms have been shown to contain proteins that will bind to Z-DNA.
台大農藝系 遺傳學 601 20000 Chapter 2 slide 34
The Organization of DNA in Chromosomes
• 1. Cellular DNA is organized into chromosomes.
A genome is the chromosome or set of
chromosomes that contains all the DNA of an organism.
• 2. In prokaryotes the genome is usually a single
circular chromsome. In eukaryotes, the genome is one complete haploid set of nuclear
chromosomes; mitochondrial and chloroplast DNA are not included.
台大農藝系 遺傳學 601 20000 Chapter 2 slide 36
Viral Chromosomes
• 1. A virus is nucleic acid surrounded by a protein
coat. The nucleic acid may be dsDNA, ssDNA, dsRNA or ssRNA, and it may be linear or
circular, a single molecule or several segments.
• 2. Bacteriophages are viruses that infect bacteria.
Three different types that infect E. coli are good examples of the variety of chromosome structure found in viruses.
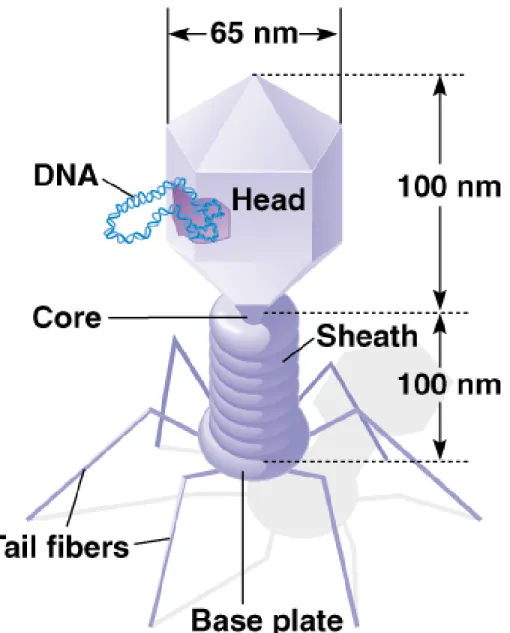
T-even phage
• 3. The T-even phages (T2, T4 and T6) have
similar structures (Figure 2.4); all have dsDNA
genomes composed of a one linear DNA molecule surrounded by a protein coat. The genomes of
these viruses show circular permutation, meaning that the starting point of each is different, because these viruses package slightly more than 100% of a genome length of DNA when they form.
台大農藝系 遺傳學 601 20000 Chapter 2 slide 38
Peter J. Russell, iGenetics: Copyright © Pearson Education, Inc., publishing as Benjamin Cummings.
Fig. 2.17 Circularly permuted and terminally redundant double-stranded DNA molecules
台大農藝系 遺傳學 601 20000 Chapter 2 slide 40
Peter J. Russell, iGenetics: Copyright © Pearson Education, Inc., publishing as Benjamin Cummings.
Fig. 2.17 Circularly permuted and terminally redundant double-stranded DNA molecules
台大農藝系 遺傳學 601 20000 Chapter 2 slide 42
ΦX174 phage
• 4. ΦX174 is a small, simple virus with one short
ssDNA chromosome
• In 1959, Robert Sinsheimer found that the DNA
of ΦX174 has a base composition that does not fit the complementary base-pair-rules. single
台大農藝系 遺傳學 601 20000 Chapter 2 slide 44
λ phage
• Bacteriophage λ is somewhat like the T-even
phages in structure. However, its chromosome changes form. A linear molecule of dsDNA is packaged inside the protein head (Fig. 2.19), but after the virus infects its host the chromosome becomes circular due to base-pairing of
complementary 12-base single-stranded regions at the ends of the linear molecule (Fig. 2.20).
台大農藝系 遺傳學 601 20000 Chapter 2 slide 46
Peter J. Russell, iGenetics: Copyright © Pearson Education, Inc., publishing as Benjamin Cummings.
Prokaryotic Chromosomes
• 1. The typical prokaryotic genome is one circular dsDNA
chromosome, but some prokaryotes are more exotic, with a main chromosome and one or more smaller ones. When a minor chromosome is dispensable to the life of the cell, it is called a plasmid. Some examples:
• a. Borrelia burgdorferi (Lyme disease in humans) has a 0.91-Mb linear chromosome, plus an additional 0.53-Mb of DNA in 17 different linear and circular molecules.
• b. Agrobacterium tumefaciens (crown gall disease of plants) has a 3.0-Mb circular chromosome and a 2.1-Mb linear one.
台大農藝系 遺傳學 601 20000 Chapter 2 slide 48
• 2. Archaebacteria also vary in chromosomal
organization, but only circular forms have been found. Examples:
• a. Methanococcus jannaschii has three chromosomes of 1.66-Mb, 58-kb and 16-kb.
• b. Archaeoglobus fulgidus has one 2.2-Mb circular chromosome.
• 3. Both Eubacteria and Archaebacteria lack a
membrane-bounded nucleus, hence their classification as prokaryotes. Their DNA is densely arranged in a cytoplasmic region called the nucleoid.
DNA Supercoiling
• 4. In an experiment where E. coli is gently lysed,
it releases one 4.6-Mb circular chromosome, highly supercoiled. A 4.6-Mb double helix is
about 1 mm in length, about 103 times longer than an E. coli cell. DNA supercoiling helps it fit into the cell.
台大農藝系 遺傳學 601 20000 Chapter 2 slide 50
Peter J. Russell, iGenetics: Copyright © Pearson Education, Inc., publishing as Benjamin Cummings.
• 5. Both positive and negative supercoiling will
condense DNA.
• 6. All organisms contain topoisomerase enzymes to supercoil their DNA.
• 7. Prokaryotes also organize their DNA into looped
domains, with the ends of the domains held so that each is supercoiled independently (Figure 2.23). The compaction factor for looped domains is about 10-fold.
• 8. The number of looped domains is species-specific
and depends on genome size. In E. coli there are about 100 domains of about 40 kb each.
台大農藝系 遺傳學 601 20000 Chapter 2 slide 52
Peter J. Russell, iGenetics: Copyright © Pearson Education, Inc., publishing as Benjamin Cummings.
Eukaryotic Chromosomes
• 1. The genome of most prokaryotes consists of
one chromosome, while most eukaryotes have a diploid number of chromosomes.
• 2. A genome is the information in one complete
haploid chromosome set. The total amount of DNA in the haploid genome of a species is its C value (Table 2.4). The structural complexity and the C value of an organism are not related,
台大農藝系 遺傳學 601 20000 Chapter 2 slide 54
• 3. Eukaryotic chromosomes are linear dsDNA,
and by weight contain about twice as much
protein as DNA. The DNA-protein complex is
called chromatin, and it is highly conserved in all eukaryotes.
台大農藝系 遺傳學 601 20000 Chapter 2 slide 56
Chromatin Structure
• 1. Both histones and non-histones are involved in physical structure of the chromosome.
• 2. Histones are abundant, small proteins with a net (+)
charge. The five main types are H1, H2A, H2B, H3 and H4. By weight, chromosomes have equal amounts of DNA and histones.
• 3. Histones are highly conserved between species (H1
less than the others).
• 4. Non-histone is a general name for other proteins associated with DNA. This is a big group, with some structural proteins, and some that bind only transiently. Non-histone proteins vary widely, even in different cells from the same organism. Most have a net (-) charge, and bind by attaching to histones. HMG (high mobility group) proteins are a well-studied example of non-histone
• 5. Chromatin formation involves histones, and
condenses the DNA so it will fit into the cell. Chromatin formation has two components:
• a. Two molecules each of histones H2A, H2B,
H3 and H4 associate to form a nucleosome core, and DNA wraps around it 1 3⁄4 times for a 7-fold condensation factor. Nucleosome cores are about 11 nm in diameter (Figures 2.25 and 2.27).
台大農藝系 遺傳學 601 20000 Chapter 2 slide 58
Peter J. Russell, iGenetics: Copyright © Pearson Education, Inc., publishing as Benjamin Cummings.
• b. H1 serves as the linker histone, connecting
nucleosomes to create chromatin with a diameter of 30 nm, for an additional 6-fold
• condensation. The exact mechanism used by H1 is
台大農藝系 遺傳學 601 20000 Chapter 2 slide 60
Peter J. Russell, iGenetics: Copyright © Pearson Education, Inc., publishing as Benjamin Cummings.
Fig. 2.26 Nucleosomes connected together by linker DNA and H1 histone to produce
台大農藝系 遺傳學 601 20000 Chapter 2 slide 62
• 6. Chromatin is arranged in looped domains of
DNA similar to those formed in prokaryotic
chromosomes. Loops are anchored to the nuclear matrix at DNA sequences called MARs (matrix attachment regions). An average human
chromosome has about 2,000 looped domains. Looped domains may be important in regulating transcription and replication (Fig. 2.30 and
Fig. 2.29 Model for the organization of 30-nm chromatin fiber into looped domains
台大農藝系 遺傳學 601 20000 Chapter 2 slide 64
Peter J. Russell, iGenetics: Copyright © Pearson Education, Inc., publishing as Benjamin Cummings.
Fig. 2.31 The many different orders of chromatin packing that give rise to the highly
Euchromatin and Heterochromatin
• 1. The cell cycle affects DNA packing, with DNA condensing for
mitosis and meiosis, and decondensing during interphase.
Chromosomes are most condensed at metaphase, when the looped domains are further coiled and the chromatic has a diameter of about 700 nm. Non-histone proteins form the scaffold for this additional condensation.
• 2. Staining of chromatin reveals two forms:
• a. Euchromatin condenses and decondenses with the cell cycle. It is actively transcribed, and lacks repetitive sequences. Euchromatin accounts for most of the genome in active cells.
• b. Heterochromatin remains condensed throughout the cell cycle. It replicates later than euchromatin, and is transcriptionally inactive. There are two types based on activity:
• i. Constituitive heterochromatin occurs at the same sites in both homologous chromosomes of a pair, and is mostly repetitive DNA (e.g., centromeres).
台大農藝系 遺傳學 601 20000 Chapter 2 slide 66
Centromeric and Telomeric DNA
• 1. Centromeres and telomeres are eukaryotic
chromosomal regions with special functions.
• 2. Centromeres are the site of the kinetochore, where
spindle fibers attach during mitosis and meiosis. They are required for accurate segregation of chromatids.
• 3. Yeast (Saccharomyces cerevisiae) centromeres are
well-studied. Called CEN regions, their sequence and organization are similar, but not identical, between the chromosomes. Other eukaryotes have different
centromere sequences, so while function is conserved, it is not due to a single type of DNA sequence. (Fig. 2-32)
Fig. 2.32 Consensus sequence for centromeres of the yeast
台大農藝系 遺傳學 601 20000 Chapter 2 slide 68
• Proteins interact with the centromere and the
spindle mircrotubule to form the kinetchore structure (Fig. 2-33)
Fig. 2.33 Hypothetical model for the kinetochore of yeast, showing the relationship of
台大農藝系 遺傳學 601 20000 Chapter 2 slide 70 • 4. Telomeres are needed for chromosomal replication and stability.
Generally composed of heterochromatin, they interact with both the nuclear envelope and each other. All telomeres in a species have the same sequence.
• a. Simple telomeric sequences are short, species-specific and tandemly repeated. (Examples: Tetrahymena is 59-TTGGGG-39, and human is 59-TTAGGG-39.) A new model suggests that the single-stranded end of the chromosome folds back to form a t-loop and then invades the double-stranded region to form a D-loop (Figure 2.34).
• b. Telomere-associated sequences are internal to the simple telomeric sequences. These complex repetitive sequences may extend many kb into the chromosome.
• c. Drosophila is unusual in having telomeres composed of transposons, rather than the short repeats seen in most eukaryotes.
台大農藝系 遺傳學 601 20000 Chapter 2 slide 72
Unique-Sequence and Repetitive-Sequence
DNA
• 1. Sequences vary widely in how often they occur within a genome. The categories are:
• a. Unique-sequence DNA, present in one or a few copies.
• b. Moderately repetitive DNA, present in a few to 105 copies. • c. Highly repetitive DNA, present in about 105–107 copies.
• 2. Prokaryotes have mostly unique-sequence DNA, with repeats only of sequences like rRNAs and tRNAs. Eukaryotes have a mix of unique and repetitive sequences.
• 3. Unique-sequence DNA includes most of the genes that encode
proteins, as well as other chromosomal regions. Human DNA contains about 65% unique sequences.
• 4. Repetitive-sequence DNA includes the moderately and highly
repeated sequences. They may be dispersed throughout the genome, or clustered in tandem repeats.
• 5. Dispersed repetitive sequences occur in families that have a characteristic sequence. Often the same few sequences are highly repeated, and comprise most of the dispersed repeats in the genome.
Little is known of their function, or indeed whether they actually serve a function. There are two types of interspersion patterns found in all
eukaryotic organisms:
• a. SINEs (short interspersed repeated sequences) with 100–500 bp
sequences. An example is the Alu repeats found in some primates,
including humans, where these 200–300 bp repeats make up 9% of the genome.
• b. LINEs (long interspersed repeated sequences) with sequences of 5 kb
or more. The common example in mammals is LINE-1, with sequences up to 7 kb in length.
• 6. Tandemly repetitive sequences are common in eukaryotic genomes, ranging from very short (1–10 bp) sequences to genes and even longer sequences. This group includes centromere and telomere sequences, and