行政院國家科學委員會專題研究計畫成果報告
非小細胞肺癌之微衛星體複製突變與核酸修補基因(hMSH2基因及
hOGG1基因)發生基因變異之相關聯姓
Microsatellite Instability and Genetic Alteratin of Two DNA Repair Genes –
hMSH2 Gene and hOGG1 Gene in Non-Small Cell Lung Cancer
計畫編號:NSC88-2314-B-002-208
執行期限:自民國87年08月01日起至民國88年07月31日
主持人:余忠仁
執行機構及單位名稱:台大醫學院內科
計畫中文摘要
基因突變現象隨著老化過程而逐步累 積於生物體中,成為癌變之潛在原因。近 來,由於發現了核酸修補系統,對於生物體 組織基因突變之調控以及修補系統基因失 常後之癌變現象,受到了廣泛的注意。微衛 星體為散佈於基因組中反覆出現之短核酸 片段,其複製變異被認為是代表遺傳因子複 製出現錯誤之標誌。在少數針對肺癌之相關 報告中,微衛星體複製突變發生率也達 33-66%。不同之微衛星體標誌於肺癌組織之突 變率,在文獻報告之差異度很大;而且至今 亦無任何有關肺癌 MMR 基因變異之相關 研究報告。我們乃提出此二年期研究計劃, 目的在評估非小細胞肺癌之微衛星體複製 突變與兩個核酸修補基因:hMSH2及 hOGG1(為新近發現,修復核酸氧化傷害之 基因),發生基因變異之相關聯性。本計劃 之第一年已收集 60 對肺癌及其非肺癌肺組 織檢體,抽取 DNA 及 RNA,並完成部分微 衛星體標誌變異之分析,目前正進行檢測核 酸修補基因在發生微衛星體複製變異之肺 癌檢體中之可能基因變異。關鍵詞:微衛星 體、基因突變、核酸錯配修補、核酸氧化傷 害、hMSH2、hOGG1、肺癌計畫英文摘要
Mutation accumulates throughout life and is the underlying cause of cancer.
Recently, much attention has been paid to the important role of the DNA mismatch repair system in controlling the accumulation of somatic mutations in human tissues and the association of mismatch repair deficiency with carcinogenesis. Microsatellite instability, representing mutations of short repetitive DNA sequence distributed within genomes, appears to be a novel mechanism in
carcinogenesis and is thought to reflect multiple replication errors from MMR genes. Frequent (33-66%) presence of replication error phenomenon in lung cancer have been reported. There were significant differences exists in the frequency of alterations in individual marker, and data regarding the alteration of MMR genes in lung cancer are lacking. We propose a two-year study to evaluate the frequency of microsatellite instability in non-small cell lung cancer, and its association with genetic alteration of MMR genes During the first year of study, we collected 60 pairs of tissues, performed microsatellite of several markers in part of the
samples, and obtained some findings. Analysis of MMR genes is undergoing. Keywords:microsatellite, mutation, mismatch repair, oxidative DNA damage, hMSH2, hOGG1, lung cancer
INTRODUCTION
Malignant tumors develop in humans for the most part in individuals of 55 years of age or more, and consist of enormous cloning expansion of cells that have accumulated mutations in defined sets of genes [1]. Mutation accumulates throughout life and is the underlying causes of cancer [2].
Statistically, it has been estimated that tumors require four to seven mutations to develop. Ionizing radiation, viral, chemical, or other physical factors are causative events to DNA mutations and are also risk factors for carcinogenesis.
Microsatellites are short (1-4 bp), repetitive, non-codifying, and highly polymorphic DNA sequences, distributed within the genome [3]. Approximately 100,000 microsatellites repeats are scattered throughout human genome. Instability of microsatellite sequence reflects malfunction in the replication or repair of DNA, so called replication error (RER) phenomenon [4]. RER can be witnessed as a change in the length of microsatellite sequences (expansions or contractions) in tumor DNA compared with constitutional DNA, but also as the complete loss of one or both alleles of the repeat locus (LOH). Microsatellite instability serves as a useful marker of a "mutator" phenotype
characteristic of hereditary nonpolyposis colorectal cancer (HNPCC) and sporadic colorectal cancers [5,6]. About 90% of HNPCC tumors had microsatellite instability, and approximately 15% of apparently
sporadic colorectal carcinomas and other types of cancers also show this abnormality [7,8]. RERs are not uncommon in lung cancer. But different from those found in colon cancer, the RERs occur in few loci, mostly localized on chromosome 3p, but can also be found on chromosome 2p, 9p and 1p [9-11].
The existence of bacteria mismatch repair (MMR) enzymes has been known for decades [12]. In Escherichia coli, HLS (mutH, mutL, mutS) system provide main repair pathways for DNA replication error. The human MMR system is believed to operate in similar fashion. The mismatch binding factor (like mutS in bacteria) includes, MSH2 and G/T mismatch binding protein (GTBP). There are at least 16 mutL-like proteins in human, MLH1, PMS1-8, PMSR1-7, showing a high genetic redundancy. No human counterpart protein for mutH has been identified so far [13]. Tumors from HNPCC patients showed inactivation of one of four MMR genes, MLH1, MSH2, PMS1, PMS2.
Apart from being responsible for the correction of biosynthetic errors in newly synthesized DNA, the MMR system is also presumed to repair exogenous chemical damage, such as lesions caused by oxidation. Oxidative damaged DNA product, 8-oxo-7,8-dihydro-guanine (8-oxoG) has gained greatest
current interest. Evidence also suggests the critical role of 8-oxoG in mutagenesis and carcinogenesis [13]. HOGG1 gene, mapped to chromosome 3p25, a region with high
frequency of RER in lung cancer, is the repair enzyme that mediate the removal of 8-oxo-G oxidative damages from DNA [14,15]
So far, only few studies concerning microsatellite instability in lung cancer have been reported. These studies demonstrated frequent RER in lung cancer. However, data regarding the alteration of MMR genes in lung cancer are still lacking. This two-year study was proposed to evaluate the frequency of microsatellite instability in non-small cell lung cancer, and the association between RER and genetic alteration of a MMR prototype protein (hMSH2) and hOGG1.
MATERIALS AND METHODS Patients and tumor tissues
Surgical specimens of tumors and the adjacent uninvolved lung tissue will be obtained from 120 patients at the time of resection. All patients should have NSCLC confirmed by histological diagnosis.
DNA and RNA extr action
Genomic DNA and total cellular RNA are extracted from tissues using the phenol-chloroform and guanidinium thiocyanate-phenol-chloroform extraction method, respectively.
Micr osatellite analysis
Microsatellite sequences are easy to
assay using PCR. Microsatellite markers for the analysis for each sample will be obtained as MAPPAIRS (Research Genetics, Huntsville, AL, USA): D2S136 (2p14-p13), D2S162 (2p25-p22) and D2S391 (2p15) on
chromosome 2p, and D3S1284 (3p13-p14), D3S1289 p14.3), D3S1067 (3p21.1-14.3), D3S1038 (3p25) and D3S1611 (3p21.3) on chromosome 3 p; IFNA and D9S171 on chromosome 9p, as well as two
monocucleotide markers: BAT-40 (1q13.1) and BAT-26 (2p). PCR will be performed by 35 cycles of amplification in a final volume of 25 µl. The PCR products are electrophoresed on 8% polyacrylamide gels containing 8M urea, dried at 80℃, and exposed to X-ray film from 24 to 72 h. The band pattern will be compared between tumorous and
non-tumorous tissues for each patient. The RER (+) tumors were defined by the presence of
microsatellite alterations in at least two different loci.
Rever se-tr anscr iption polymer ase chain r eaction of hMSH2 and hOGG1
RNA of tissues obtained from those patients with microsatellite instability will be used to determine hMSH2 and hOGG alteration. Two µg of total RNA is reverse transcribedand subjecterd to PCR
amplifications in 50 µl reactions.
Sequence analysis of mutation
The PCR products are purified by GENECLEAN III(Bio101, Vista, CA, USA) and subjected to automatic sequencing. DNA sequencing is performed by an Applied
Biosystem 373A DNA sequencer (Foster City, CA, USA) using the dye-terminator method. Putative mutations are confirmed with a second round of sequencing reaction.
Immunohistochemistr y
Antibodies of different MMR proteins (hPMS1, hPMS2, hMLH1, hMSH2, GTBP) will be applied. The staining pattern of MMR proteins are normally nuclear. Tumor cells that exhibit an absence of nuclear staining in the presence of non-neoplastic cells with nuclear staining will be considered to have an abnormal pattern.
RESULTS
After evaluating several microsatellite markers in 12 pairs of lung cancers , the results showed that : 4 mutants out of 12 were detected in BAT-40 (Fig. 1), 2/12 in BAT-26, and 1/12 in D2S136, while none in D2S162. Fig. 1 shows LOH of BAT-40 in T222, and insertion in T225
The PCR products for hOGG1 and hMSH2 were generated with the fragments size described as following:
Fig.2 PCR products of hOGG1exon 1-7
◎
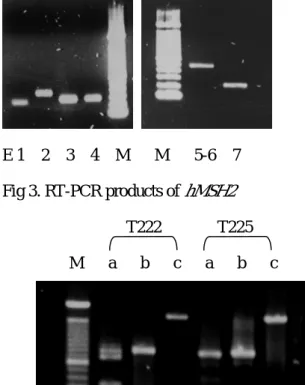
E 1 2 3 4 M M 5-6 7 Fig 3. RT-PCR products of hMSH2
T222 T225 M a b c a b c
a: codon 1-247, b: codon 241-507, c: codon 381-934.
REFERENCES
1. Fearson ER and Vogelstein B. Cell 1990;61:759-67.
2. Vogelstein B and Kinzler KW. Trends Genet 1993;9:138:41.
3. Ramel C. Environ Health Prospect 1997;105 (suppl 4):781-9.
4. Ionov Y, Peinado MA, Malkhosyan S, Shibata D, and Perucho M. Nature 1993; 363:558-61.
5. Liu B, Farrington SM, Petersen GM, et al. Nature Med. 1995;1:348-52.
6. Liu B, Parson R, Papadopoulos N, et al. Nature Med 1996;2:169-74.
7. Keller G, Grimm V, Vogelsang H, et al. Int J Cancer 1996;68:571-6.
8. Shridhar V, Siegfried J, Hunt J, Alonso MM, and Smith DI. Cancer Res 1994;54:2084-7.
9. Merlo A, Mabry M, Gabrielson E, Vollmer R, Baylin SB, and Sidransky D. Cancer Res 1994;54:2098-101.
10. Fong KM, Zimmerman PV and Smith PJ. Cancer Res 1995;55:28-30.
11. Su SS, Modrich P. Proc Natl Acad Sci USA 1986;83:5057-61.
12. Fishel R and Wilson T. Curr Opin Genet Develop 1997;7:105-13.
13. Grollman AP and Moriya M. Trends Genet 1993;9:246-9.
14. Van der Kemp PA, Thomas D, Barbey R, de Oliveira R, and Boiteux S. Proc Natl Acad Sci 1996;93:5197-202.
15. Radicella JP, Dherin C, Desmaze C, Fox MS, and Bioteux S. Proc Natl Acad Sci USA 1997;94:8010-5.