National Chiao-Tung University PhD Dissertation 國立交通大學博士學位論文
Prevalence Analysis and Expressional Control of
Klebsiella pneumoniae Fimbriae
克雷白氏肺炎桿菌線毛的分佈與表現
生物科技學系 博士班 學生:吳健誠(9228509) Student:Chien-Chen Wu 指導教授:彭慧玲博士 Advisor:Hwei-Ling Peng, Ph. D. 中華民國九十九年十二月 December, 2010謝誌 (Acknowledgement) 1999 年,我離開從小居住的台北來到新竹,轉眼間已經過了 11 年了。現在回想 起這一段日子,許多經過的事、遇見的人,心裡有感慨萬千,要感謝的人也實在太 多。在這本論文完成同時,我想起大二升大三的暑假,我進入了實驗室進行專題研 究,那是我大學生活的轉捩點,而後就這麼一路沒有間斷地直到現在。在這段過程 中,首先我要感謝我的指導教授彭慧玲老師,給予我機會在實驗室進行研究。彭老 師總是充滿耐性、細心地教導並且關心我的感受。我還記得當我是專題生時,老師 每週都會撥空與我討論,從很基本的實驗細節與原理、學術論文的撰寫,以至於研 究方向的邏輯,可以說是一點一滴、一字一句地指導我。彭老師總是給予學生很大 的空間自由發展與嘗試。而開始專題研究後,過去認為枯燥無聊的教科書內容,變 成了實際應用於研究的原理基礎,讓我重拾學習的興趣,可以說改變了我的人生。 而在學術以外,我也要謝謝彭老師一直以來包容我個性上的缺點與不成熟,讓我有 機會反省、學習與成長。 感謝清華大學的張晃猷老師,對於我的研究總是能給予最精闢的建議。張老師 在學術上不懈的熱情以及廣博的知識,一直是我的學習榜樣。在論文以及研究計畫 的撰寫上更曾經專門開課指導我們,令我獲益良多。每次開會討論時,張老師獨特 的見解常常讓我注意到自己邏輯上的缺失、讓我深思並且學習到很多。張老師舉辦 的多次聚會與出遊,也成為我非常美好的回憶。同時也感謝張老師實驗室的成員們, 特別是韻如與莉芳學姐。 感謝林志生老師從大學以來各方面的指導與關心。林老師在教學以及研究上認 真嚴謹的態度是我學習的模範。在林老師的課堂上,不僅能得到充實的專業知識, 也能學習到科學家應該具備的能力與態度。每次林老師見到我時,都會關心我的狀 況、給予我人生及學業上的建議,我能感受到老師對於學生們的熱情與期許。謝謝 老師,我會繼續努力不讓老師失望。 感謝楊昀良老師一直以來親切地給予我指導與建議,常常以不同的角度切入我 的研究,提出我從沒想過的重要問題。楊老師總是能以有趣輕鬆的方式教授複雜的 學問,讓我們在學習時能自由思考不受侷限。老師和善的態度,也讓我在許多次口 試時得以減輕緊張,謝謝老師。 感謝中興大學的鄧文玲老師,在專業領域上給予我非常多的幫助、解決我實驗 上的困難。鄧老師在學術上的熱忱與豐富知識,讓我在每次談話後都收穫豐富,並 且對於自己的研究更有動力。鄧老師在會議討論時直接爽朗的作風,更讓我感覺到 作研究是件單純快樂的事。 感謝中山醫學大學的賴怡琪老師,在CG43 建立了良好的研究模式,精細優美的 實驗設計與論文撰寫一直是我的榜樣。在會議討論時,怡琪學姐總是溫柔親切地回 答我的問題,也感謝學姐在我最低潮的時候曾為我加油打氣。
感謝清華大學的游萃蓉老師,以及其實驗室的安琪、國良、義洋。多次在會議 中的接觸以及合作進行研究時,都讓我深自檢討並獲益良多。游老師積極進取、有 效率的做事態度與方法,是我非常佩服與效法的模範。 感謝彭老師實驗室眾多的學長姐們。首先是在我初進實驗室時,可愛的巧韻學 姐盡心盡力並且嚴格地帶領我入門。感謝丸子學姐與我一起在線毛的領域並肩奮鬥 了多年、靖婷學姐在我實驗瓶頸時總是能讓我重燃希望動力,兩位大學姐營造了快 樂的實驗室氣氛,讓我擁有許多珍貴的回憶。感謝學識淵博的盈璁學長,常讓我發 覺自己的不足之處。感謝學長姐們:沈惠、騰逸、怡欣、致翔、美甄、定宇、珮瑄、 平輝、婉君。當小學弟的日子是我最快樂也最懷念的時期,非常慶幸能夠有你們的 陪伴。 感謝與我一起走過博士班每個階段的小新,分擔了一路上許多的壓力痛苦,常 常與我激烈地討論實驗,或著笑鬧分享生活點滴。感謝祐俊,在討論時常有令我驚 嘆的見解,並且開啟了我們實驗室c-di-GMP的研究。也感謝所有交大生科 92 級的同 學們,特別是少偉、愛雁、頌瑾、怡伶、小操、繼元、隆勇、dede、柳李、琮道、淑 惠、青峰、晞均、羿喬、阿伯、小天,這麼多年來與我一起苦讀、玩樂、奮鬥,不 論在我快樂或痛苦時,總是能與你們分享、一起走過。 感謝靜柔,在我博士班後期成為實驗室的支柱,減輕我許多壓力,常常陪我吃 飯、喝咖啡,在聊天中排解我許多的困擾。感謝實驗室的學弟妹們,特別是育聖、智 凱、格維、朝陽、登魁、秉熹、大大、fish、雅雯、顗峰、朝彥、信文、崴云、品瑄 ,與我共同度過、容忍我作為學長的不成熟之處,在相處中也讓我學習到很多。 雖然實驗室生活有許多愉快的回憶,但在過程中仍然有著低潮與困惑,以及大 部分時候不被瞭解的痛苦。因此我要感謝在實驗室以外,支持我度過這一段日子的 好朋友們,在每次的聚會後讓我能重新振作回到實驗室繼續努力!感謝陶藝社以及 應用藝術研究所的朋友們:昱佑、Vic、彥均、宗凰、庭毓、陳小新、劭恆、老羅、志 祥、明鴻、小宏、小黃、莎莎,懷念那些伴隨著吉他與歌聲的美麗夜晚。感謝高中 同學們,特別是伯鑫、彭跟小胖。感謝朝陽15th的朋友們:邦桓、明玲、微東、聖慈 、騷、文宣、維秀、阿潔,一直期待我能趕快畢業回去台北。感謝與我住了六年的 室友建賢,忍受我常常半夜才回到寢室時發出的噪音,與我一起睡前閱讀或徹夜長 談,如同兄長般地照顧我的生活。 最後要感謝我的爸媽,從小重視我的教育,盡全力給我最好的成長與學習環境、 以及無條件的愛與關心。也要謝謝三位姐姐跟哥哥,在我求學期間照顧媽媽。由於 有家人在背後的支持,才讓我能自由地作我想作的事。 感謝所有曾參與我生命的你們,豐富並成就了現在的我。因為有你們的陪伴, 令我能微笑著回想過往的點點滴滴。
論文摘要 克雷白氏肺炎桿菌是一株引起伺機性感染的革蘭式陰性細菌,大多於免疫力 不全的病人身上,引起化膿性的潰瘍、菌血症、尿道以及呼吸道的感染。為了探 討克雷白氏肺炎桿菌的致病機制,在本論文中,我們針對在感染初期扮演著重要 角色的線毛的分佈情況及表現調控進行研究。 我們藉由分析克雷白氏肺炎桿菌 NTUH-K2044 的全基因體序列,共發現了 九組獨立的線毛基因群。除了過去已知的第一型與第三型線毛基因組外,其餘的 七組尚未有文獻報導,我們將其分別命名為kpa、kpb、kpc、kpd、kpe、kpf、kpg 基因組。我們進一步分析了克雷白氏肺炎桿菌的臨床分離菌株,發現kpb 與 kpc 基因組在K1 莢膜血清型的菌株中有顯著較高的存在率。隨後,我們針對 Kpc 線 毛進行了特性分析。在大腸桿菌中表現Kpc 線毛的生合成基因組 kpcABCD 後, 可以致使細菌製造出 Kpc 線毛並且增強其生物膜的形成能力。我們也發現 Kpc
線毛的表現受到了相變機制(phase variation)的調控,而此相變機制則是由 KpcI
專一性DNA 重組酶所負責。最後,我們著重於第三型線毛的表現調控進行研究。 我們發現位於第三型線毛基因組下游的mrkHIJ 操作組可生合成出三個調控蛋白 質來影響第三型線毛的表現。mrkI 基因被預測可生合成出 LuxR 類型的轉錄因 子,而我們將 mrkI 在克雷白氏肺炎桿菌中進行剔除後,發現會破壞第三型線毛 的基因轉錄。同時我們也證明,具有PilZ 模組的 MrkH 與具有 EAL 模組的 MrkJ, 可分別正向與負向調控第三型線毛的表現。此外,我們發現Fur 攝鐵調控子會經
由調控 mrkHIJ 操作組來影響第三型線毛的表現。而二次代謝傳導物 c-di-GMP 也被證明可經由Fur 與 MrkI 來活化第三型線毛的表現。我們還發現缺氧環境可 能是影響第三型線毛表現的因子。 在本論文中,我們首度報導了克雷白氏肺炎桿菌線毛的基因體學分析,這個 報導將有助於未來對於此細菌線毛黏附作用的研究(第二章)。我們也分析了 Kpc 線毛的功能性與相變調控表現(第三章),以及探討影響第三型線毛表現的 多重調控因子(第四章)。
Thesis Abstract
Klebsiella pneumoniae is a Gram-negative pathogen which causes suppurative
lesions, bacteremia and urinary as well as respiratory tract infections mostly in patients with underlying diseases. To investigate the pathogenicity of K. pneumoniae, we focused on the study of prevalence and expression of fimbriae, which are important virulence determinants during the initial infection.
By analyzing the whole genome sequence of K. pneumoniae NTUH-K2044, nine distinct fimbrial gene clusters were identified. Besides type 1 and type 3 fimbrial genes, the other seven are novel and designated kpa, kpb, kpc, kpd, kpe, kpf, and kpg. The following prevalence analysis among K. pneumoniae clinical isolates indicated that the kpb and kpc genes were more prevalent in the strains of capsular serotype K1. Subsequently, the Kpc fimbria, encoded by kpcABCD genes, was characterized. Induced expression of the recombinant kpcABCD genes in Escherichia coli resulted in Kpc fimbriation and increased bacterial biofilm formation. The Kpc fimbriae expression was also found to be regulated under phase variation mediated by the site-specific recombinase KpcI. Finally, the expressional control of type 3 fimbriae in
K. pneumoniae CG43 was investigated. We described that the type 3 fimbriae
expression was mediated by three regulatory proteins encoded by the mrkHIJ operon which located downstream to the type 3 fimbrial genes. Deletion of mrkI, which
encoded a LuxR-type response regulator, from K. pneumoniae was found to abolish the expression of type 3 fimbriae at transcriptional level. Moreover, MrkH, a PilZ domain protein, and MrkJ, an EAL domain protein, appeared to act as an activator and a repressor, respectively, for the type 3 fimbriae expression. Besides, we showed that the ferric uptake regulator Fur could activate the expression of type 3 fimbriae through regulation on the mrkHIJ operon. The second messenger c-di-GMP was also found to activate the expression of type 3 fimbriae through Fur and MrkI. Furthermore, we identified that oxygen-limitation was possibly an environmental stimulus for activating the type 3 fimbriae expression.
In this dissertation, we reported the first genomic analysis of fimbrial gene sequences in K. pneumoniae, which pave the way for future study of the bacterial adherence (Chapter 2). Subsequently, the functional role and phase-variable expression of Kpc fimbriae were characterized (Chapter 3). Moreover, a multi-factorial regulation of type 3 fimbriae expression was elucidated (Chapter 4).
Table of Contents
謝誌 (Acknowledgement) ...I 論文摘要 (Thesis Abstract in Chinese) ...III Thesis Abstract ...V Table of Contents ...VII List of Tables ...X List of Figures ...XI Abbreviations ...XIII Chapter 1.
General Introduction ...1
1.1. Klebsiella pneumoniae ...2
1.1.1. K. pneumoniae infections ...2
1.1.2. K. pneumoniae virulence factors ...4
1.2. Fimbriae (Pili) ...7
1.2.1. K. pneumoniae type 1 fimbriae ...9
1.2.2. K. pneumoniae type 3 fimbriae ...11
1.3. Cyclic-di-GMP signaling ...13
1.4. Thesis objectives ...16
Chapter 2. Prevalence Analysis of the Fimbrial Gene Clusters in Klebsiella pneumoniae...20
2.1. Abstract ...21
2.2. Introduction ...22
2.3. Results ...24
2.3.1. Identification of the fimbrial gene clusters in K. pneumoniae NTUH-K2044 ...24
2.3.2. PCR screening for the presence of the fimbrial genes ...25
2.3.3. PCR-RFLP analysis of the mrkD genes ...26
2.4. Discussion ...27
Chapter 3. Regulation of Kpc Fimbriae by the Site-specific Recombinase KpcI...34
3.1. Abstract ...35
3.2. Introduction ...36
3.3. Results ...38
3.3.3. The KpcI recombinase is probably the regulator for the Kpc fimbriae ...40
3.3.4. Expression of kpcI leads to inversion of kpcS ...41
3.3.5. Effect of the recombinant KpcI196 on the switching of kpcS ...42
3.3.6. KpcI-mediated expression of KpcA ...43
3.3.7. The transcription of Kpc fimbrial genes is impeded in the kpcS-ON K. pneumoniae cells ...44
3.4. Discussion ...46
Chapter 4. Regulation of the Expression of Type 3 Fimbriae in Klebsiella pneumoniae CG43...62
4.1. Abstract ...63
4.2. Introduction ...65
4.3. Results ...68
4.3.1. mrkHIJ is transcribed in a polycistronic mRNA ...68
4.3.2. Deletion of mrkI represses the expression of type 3 fimbriae ...68
4.3.3. Identification of the transcription start site of mrkA ...69
4.3.4. Deletion of mrkI decreases the transcription of mrkA ...70
4.3.5. Activity of MrkI is probably affected by phosphorylation ...70
4.3.6. Overproduction of MrkH increases the expression of type 3 fimbriae ...72
4.3.7. Deletion of fur represses the expression of type 3 fimbriae ...73
4.3.8. Deletion of fur and mrkI represses the activity of PmrkH ...74
4.3.9. Extracellular iron availability affects the expression of type 3 fimbriae ...75
4.3.10. Fur and MrkI are required for the c-di-GMP-activated expression of type 3 fimbriae ...76
4.3.11. Knockout of fur and mrkI decreases K. pneumoniae biofilm formation ...77
4.3.12. The expression of type 3 fimbriae under the oxygen-limiting conditions ...78
4.4. Discussion ...81
Chapter 5. Conclusion and Perspectives ...107
Chapter 6.
Experimental Sections ...112
6.1. Materials ...113
6.1.1. Plasmids, primers, bacterial strains and growth conditions ...113
6.2 General Experimental Procedures ...114
6.2.1. Bioinformatics ...114
6.2.2. PCR detection of fimbrial genes ...114
6.2.3. KpcA antiserum preparation ...115
6.2.4. Construction of the expression plasmid pETQ ...115
6.2.5. Construction of fimbriae expression plasmids ...115
6.2.6. Transmission electron microscopy (TEM) ...116
6.2.7. Immunofluorescence microscopy analysis ...116
6.2.8. Biofilm formation assay ...117
6.2.9. Yeast-cell agglutination (YA) ...117
6.2.10. Switch orientation assay ...118
6.2.11. Construction of specific gene-deletion in K. pneumoniae NTUH-K2044 ...118
6.2.12. Construction of specific gene-deletion in K. pneumoniae CG43 ...119
6.2.13. Construction of the reporter fusion plasmids and measurement of promoter activity ...121
6.2.14. Identification of the operon structure by reverse-transcription PCR (RT-PCR) ...122
6.2.15. Identification of mrkA transcriptional start site ...123
6.2.16. Construction and expression of the recombinant proteins ...124
6.2.17. Construction of the site-directed mutants derived from K. pneumoniae CG43S3 ...126 6.2.18. Statistical methods ...126 References...137 Publication ...165 Vita ...166
List of Tables
Table 2.1. Prevalence of kpb and kpc genes in K. pneumoniae isolates ...29
Table 2.2. Repertoire of fimbrial genes among K. pneumoniae isolates with different K serotypes ...30
Table 2.3. K serotypes of the K. pneumoniae clinical isolates ...31
Table 6.1. Bacterial strains used in this study ...128
Table 6.2. Plasmids used in this study ...130
List of Figures
Fig. 1.1. Domain architecture of putative c-di-GMP signaling proteins encoded by
the K. pneumoniae NTUH-K2044 genome ...18
Fig. 2.1. Fimbrial gene clusters of the chaperone-usher-dependent assembly class in K. pneumoniae NTUH-K2044 ...32
Fig. 2.2. PCR amplicons of the pilin and adhesin encoding genes in K. pneumoniae NTUH-K2044 ...33
Fig. 3.1. Transmission electron micrographs of recombinant Kpc fimbriae ...49
Fig. 3.2. Specificity of the KpcA antiserum ...50
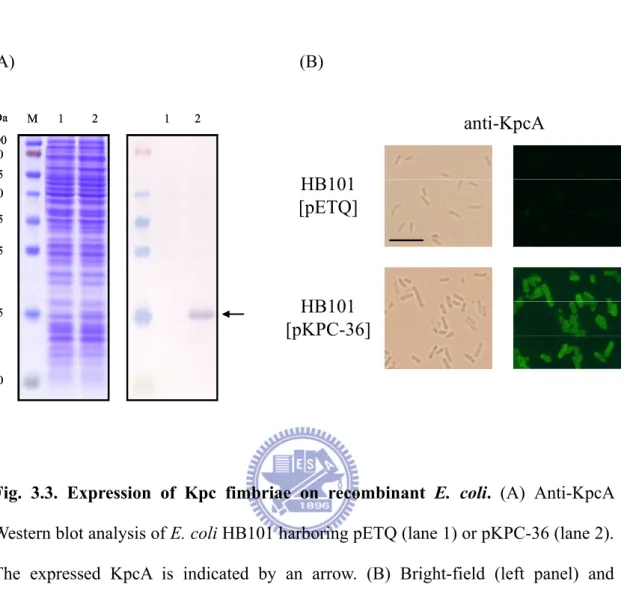
Fig. 3.3. Expression of Kpc fimbriae on recombinant E. coli ...51
Fig. 3.4. Biofilm forming ability of E. coli expressing the Kpc fimbriae ...52
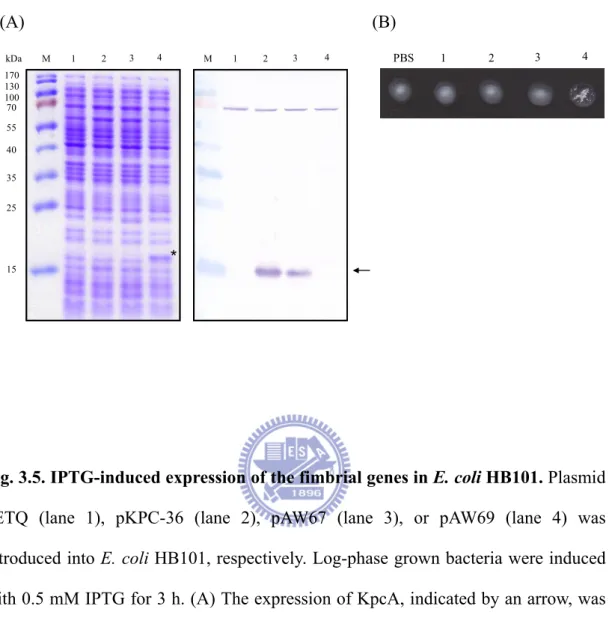
Fig. 3.5. IPTG-induced expression of the fimbrial genes in E. coli HB101 ...53
Fig. 3.6. Alignment of the amino acid sequences of the fimbrial recombinases ...54
Fig. 3.7. Sequence analysis of the putative promoter region of the kpc gene cluster ...55
Fig. 3.8. KpcI196-mediated inversion of kpcS ...56
Fig. 3.9. Determination of the promoter activities of kpcSON and kpcSOFF ...57
Fig. 3.10. The recombinant KpcI mediated the kpcS inversions in both directions ....58
Fig. 3.11. KpcI-mediated expression of KpcA in E. coli ...59
Fig. 3.12. The T5lac promoter driven expression of kpcABCD genes in K. pneumoniae ...60
Fig. 3.13. Determination of the promoter activities of kpcSON and kpcSON* ...61
Fig. 4.1. Schematic gene organization of a chromosomal region encoding K. pneumoniae type 3 and type 1 fimbriae ...90
Fig. 4.3. Deletion of mrkI decreases the expressions of type 3 fimbriae ...92
Fig. 4.4. Identification of mrkA transcription start site by 5’-RACE ...93
Fig. 4.5. Deletion of mrkI decreased the transcription of mrkA ...94
Fig. 4.6. MrkI is probably a response regulator activated by phosphorylation ...95
Fig. 4.7. Amino acid sequence alignment of PilZ domain proteins ...96
Fig. 4.8. MrkH-mediated activation of type 3 fimbail expression ...97
Fig. 4.9. Deletion of fur repressed the expression of type 3 fimbriae ...98
Fig. 4.10. The promoter activity of the upstream region of mrkH was regulated by Fur and MrkI ...99
Fig. 4.11. Extracellular iron availability affected the expression of type 3 fimbriae ... 100
Fig. 4.12. Fur and MrkI were required for the c-di-GMP-activated type 3 fimbriae expression upon YdeH overproduction...101
Fig. 4.13. Effects of mrkH deletion and overexpression on type 3 fimbriae expression ...102
Fig. 4.14. Knockout of fur, mrkI, and mrkJ decreased K. pneumoniae biofilm formation ...103
Fig. 4.15. Correlation between type 3 fimbriae expression and oxygenation ...104
Fig. 4.16. Deletion of rcsB slightly decreased the expressions of type 3 fimbriae ...105
Fig. 4.17. A model illustrating the regulation of the expression of type 3 fimbriae mediated by Fur, MrkH, MrkI, and MrkJ in K. pneumoniae CG43 ...106
Abbreviations
bp base pair(s)
CPS capsular polysaccharide CFU colony forming unit
c-di-GMP Bis-(3’-5’)-cyclic dimeric guanosine monophosphate DGC di-guanylate cyclase
EDTA ethylenediaminetetraacetic acid ESBL extended-spectrum β-lactamase
EMSA electrophoretic mobility shift assay h hour(s)
IPTG isopropyl 1-thio-β-D-galactopyranoside
kb kilobase(s) kDa kilodalton(s) LB Luria-Bertani μg microgram ml mililiter μl microliter mM milimolar μM micromolar OD optical density
ONPG ο-nitrophenyl-β-D-galactopyranoside
PAGE polyacrylamide gel electrophoresis
PDE phosphodiesterase
qRT-PCR quantitative real-time polymerase chain reaction RFLP restriction fragment length polymorphism rpm revolutions per minutes
RT-PCR reverse transcription polymerase chain reaction SDS sodium dodecyl sulfate
CHAPTER 1
1.1. Klebsiella pneumoniae
Klebsiella pneumoniae is a Gram-negative bacterium that belongs to the gamma
subdivision of the class Proteobacteria and exhibits relatively close genetic relatedness to the other genera of Enterobacteriaceae, including Escherichia,
Salmonella, Shigella, and Yersinia (20). Klebsiella spp. are ubiquitous in nature and
characterized as rod-shaped, non-motile, usually encapsulated bacteria that can live in water, soil, and plants, and infect humans and animals (243, 244). In humans, K.
pneumoniae behaves like a commensal mainly in the nasopharyngeal and intestinal
mucosae (244). In this respect, the genus Klebsiella is like Enterobacter and
Citrobacter but unlike Shigella spp. or E. coli, which are common in humans but not
in the environment (244).
1.1.1. K. pneumoniae infections
K. pneumoniae is an opportunistic pathogen frequently involved in severe
nosocomial infections in immunocompromised individuals who are hospitalized and suffer from severe underlying diseases, such as diabetes mellitus or chronic pulmonary obstruction (244). K. pneumoniae is responsible for a variety of diseases including suppurative lesions, bacteriemia, urinary tract infections, pneumonia, and sometimes life-threatening septic shock (23, 149, 165, 236, 244). The clinical pattern
of K. pneumoniae infection in humans has changed since this organism was discovered in the 1880s (97, 98). Until the 1960s, K. pneumoniae was an important cause of community-acquired pneumonia (42), however, the incidence of this type of infection has dropped, and hospital-acquired K. pneumoniae infection now predominates (105, 244, 303). Since 1980s, K. pneumoniae is emerging as an important pathogen both in the community and the hospital setting (161). In the hospital environment with the extensive use of antibiotics, multiple drug resistance has been increasingly observed in K. pneumoniae, especially the extended-spectrum β-lactamase (ESBL)-producing strains (93, 96, 161, 208, 212). Carbapenems are considered to be the preferred agents for the treatment of serious infections caused by ESBL-producing K. pneumoniae because of their high stability to β-lactamase hydrolysis and observed retained susceptibility of ESBL producers (61). However, K.
pneumoniae isolates resistant to carbapenems have been reported worldwide since
2000s (128, 169, 224, 329). The emergence of carbapenem-resistant enterobacteria is worrisome because of the option for antimicrobial treatment is further restricted.
The emergence of an invasive form of the community-acquired K. pneumoniae infection, which presents as primary bacteremic liver abscesses, endophthalmitis, and meningitis (50, 89, 90, 189, 246, 315), has been reported almost exclusively in Asia (100), especially in Taiwan (100, 302, 315). Although these invasive and highly
encapsulated K. pneumoniae strains are universally resistant to ampicillin, they are unable to produce ESBL and susceptible to most antibiotics (178, 179). In addition, approximately 50 to 75% of the patients with K. pneumoniae liver abscess also presented with diabetes mellitus (177, 217, 300). Although the preponderance of this severe invasive K. pneumoniae infection remains unknown, the involvement of both host and microbial factors during pathogenesis could be anticipated.
1.1.2. K. pneumoniae virulence factors
A number of bacterial factors that contribute to K. pneumoniae pathogenicity have been identified, which include capsular polysaccharide (CPS), lipopolysaccharide (LPS), iron acquisition systems, and adherence factors (244). Clinically isolated K. pneumoniae usually produced large amount of CPS and therefore forms large glistering colonies with viscid consistency. The abundant CPS that typically surrounds K. pneumoniae protects against the bactericidal action of serum and impairs phagocytosis (11, 63), and may be regarded as the most important virulence determinant of K. pneumoniae. Among the 77 described capsular (K) types of the serotyping scheme, serotypes K1, K2, K4 and K5 are highly virulent in experimental infection in mice and are often associated with severe infections in humans and animals (216, 220, 229, 281). Furthermore, the K1 and K2 serotypes
were found to be the most prevalent capsular serotypes in liver abscess-causing K.
pneumoniae (100, 178).
Genetic determinants for K1 and K2 CPS biosynthesis and regulation have been reported (15, 58). In K. pneumoniae NTUH-K2044 of serotype K1, deletion of the mucoviscosity associated gene A (magA) abolishes the CPS biosynthesis and thus reduces the bacterial virulence (88). The gene magA is only contained in the K1 cps gene cluster and hence could be applied to rapidly detect K. pneumoniae strains of serotype K1 (328). A PCR analysis for the K2 capsule-associated gene A (k2A) has also been used to identify K. pneumoniae strains of serotype K2 (74, 330). The presence of rmpA (regulator of the mucoid phenotype A) gene correlated with abscess formation in patients with community-acquired K. pneumoniae bacteremia and attributed to be a risk factor for metastatic infection in patients with K. pneumoniae liver abscess (180, 331). The rmpA together with rmpA2 gene both located on the large virulence plasmid pLVPK (48, 295) are able to enhance the CPS biosynthesis thereby confer K. pneumoniae a hypermucoviscosity phenotype (53, 171).
LPS comprising three parts, lipid A, core, and O antigen, is responsible for the resistance to complement-mediated killing as well as antimicrobial peptides attack, and the establishment of septic shock (7, 52, 87, 211). Antimicrobial peptides, such as
polymyxin B, are bactericidal agents that exert their effects by interacting with the LPS of Gram-negative bacteria. The polycationic peptide ring on polymyxin competes for and substitutes the calcium and magnesium bridges that stabilized LPS, thus disrupting the integrity of the outermembrane leading to cell death (118, 332). In our previous study, the genetic determinants for LPS modification and CPS level have been shown to involve in polymyxin B resistance of K. pneumoniae CG43 (52).
Iron starvation is one of the major barriers that virulent bacteria must overcome in order to proliferate in the host. Multiple iron-acquisition systems have been described in K. pneumoniae (136). Analysis of the genomic sequence of K.
pneumoniae NTUH-K2044 revealed 10 putative iron-acquisition systems, whereas K. pneumoniae strain MGH78578 and CG43 possess only 6 and 8 of these systems,
respectively (136, 185). Prevalence study and animal experiment have been performed to assess the role of iron-acquisition systems in K. pneumoniae pathogenicity (136, 220, 295). Adherence factors possessed by K. pnuemoniae including type 1 and type 3 fimbriae, which play crucial roles in adhesion to host cells, persistence, and biofilm formation, are focused in this thesis and introduced in detail below. Other K. pneumoniae virulence determinants involved in acid resistance (135), oxidative stress response (123), and allantoin metabolism (195) have also been reported.
Complete genome sequence of K. pneumoniae strains MGH78578 (230) and NTUH-K2044 (325), which are human pathogens respectively isolated from sputum and liver abscess, and a nitrogen-fixing endophyte strain 342 (95) have been determined. Genomic and phenotypic analyses have also been performed among 235
K. pneumoniae strains in order to identify the evolutionary emergence of virulent
clones (37).
1.2. Fimbriae (Pili)
Successful establishment of infection by bacterial pathogens requires adhesion to host cells, colonization of tissues, and in certain cases cellular invasions, followed by intracellular multiplication, dissemination to other tissues, or persistence (242). Fimbriae, also called pili, are hair-like appendages that extend out of the bacterial cell surface and exert on bacterial attachment and invasion, biofilm formation, cell motility and transport of proteins and DNA across membranes. Based on their biosynthetic pathway, these non-flagellar appendages of Gram-negative bacteria can be categorized into five major classes: chaperone-usher (CU) fimbriae, curli, type 4 pili, type 3 secretion needle, and type 4 secretion pili (99). Of these five classes, the CU fimbriae are the most extensively studied and often constitute important virulence factors, responsible for specific host attachment and/or the evasion of host responses
(323, 333). CU fimbriae are assembled into linear, unbranched polymers consisting of several hundreds to thousands of pilus subunits (also known as pilins) that range in size from ~12 kDa to ~20 kDa. Generally, the major pilin constitutes the fimbrial rod and an adhesin located at the tip to mediate its specific binding activity. Besides, there are minor pilins which incorporate into the fimbrial rod and affect assembly and adherence activity for several types of fimbriae (18, 139, 155, 186, 310). The machinery of fimbrial assembly is highly conserved, which comprises a periplasmic chaperone and an outer-membrane usher proteins (99, 240, 263, 265, 299, 311, 314). The fimbrial subunits are secreted by general secretion pathway into the periplasm and protected by the fimbrial chaperone from degradation by protease, and then transported to the usher for the assembly.
An early hierarchical classification of CU systems based on conserved structural elements in the chaperones identified two distinct subgroups, FGL- and FGS-chaperone assembled pili, which correspond with the assembly of thin fibrillar and rod-like pili, respectively (147). However, recent phylogenetic analysis in 189 CU systems revealed 6 main clades: α-, β-, γ- (which is subdivided into γ1, γ2, γ3, and γ4),
κ-, π- and σ-fimbriae, based on common usher ancestry, and supported by similarities in operon structure and morphology of organelles within the separate clades (226). Among these, the rod-like or typical fimbrial organelles are found in the α-, γ- and π-
fimbrial clades. These CU fimbrial genes, respectively encode pilins, chaperone, usher, and adhesin, generally transcribe in an operon manner, and multiple CU fimbrial gene clusters are often presented in a Gram-negative bacterial genome (192, 301, 305).
Most of K. pneumoniae strains possess two types of CU fimbriae, type 1 and type 3 fimbriae (belonged to the γ1 and γ4- fimbriae, respectively), which are
introduced in detail below. The afimbrial adhesin CF29K (67, 70), KPF-28 fimbriae (72), and a capsule-like extracellular afimbrial adhesin (91) have also been reported for some K. pneumoniae strains.
1.2.1. K. pneumoniae type 1 fimbriae
Type 1 fimbriae are approximately 7 nm wide and 1-2 μm long surface organelles found in virtually all members of the family Enterobacteriaceae (119, 163). They are well known for the ability to bind to mannose-containing structures on host cells and extracellular matrix. Bacteria expressing type 1 fimbriae are able to cause mannose-sensitive agglutination of yeast cells or erythrocytes (mannose-sensitive haemagglutination, MSHA) from guinea pig. Furthermore, type 1 fimbriae has been shown to play a crucial role during urinary tract infections by mediating adhesion to mannose-containing receptors on the uroepithelium and promoting the formation of intracellular bacterial communities (62, 145, 162, 174, 219, 252, 285, 290, 323).
Type 1 fimbriae have been most extensively studied in E. coli, and the corresponding structures of K. pneumoniae are highly similar with regard to genetic composition and regulation (60, 107, 267, 290). However, there are significant genetic, serological, and functional differences between type 1 fimbria variants in the different species (60, 80, 108, 197). E. coli type 1 fimbriae are encoded by the fimAICDEFGH operon. The fimbrial rod consists of the major subunits FimA and the minor subunits FimI, FimF, and FimG. The adhesive properties of type 1 fimbriae are exerted by the FimH adhesin which locates at the tips of the fimbriae. FimC and FimD are respectively chaperone and usher that are required for the fimbrial assembly. Unique to the K. pneumoniae fim gene cluster is the fimK gene, which locates downstream to the fimH gene and encodes an EAL domain protein (252). Deletion of fimK has been shown to activate the type 1 fimbriae expression in K. pneumoniae (252). In addition,
fimB and fimE genes located upstream to the fim operon encode DNA recombinases
that mediate the expression of type 1 fimbriae (4, 29, 267, 290).
In E. coli, the regulatory network of type 1 fimbriae expression is extensively studied. Environmental conditions such as osmolarity and pH are involved in the modulation of the type 1 fimbriae expression (101, 276). Second messengers cAMP and (p)ppGpp as well as regulatory proteins, including Lrp, IHF, RpoS, NanR, NagC, CRP, and H-NS, have also been described to affect the expression of type 1 fimbriae
(1, 30, 75, 82, 83, 102, 157, 160, 218, 228, 283). However, whether these regulators exert similar effects in K. pneumoniae awaits further investigation. Several reports indicated that K. pneumoniae poorly expresses type 1 fimbriae in vitro (252, 267, 290), and the expression is phase-variable (267, 290). In addition, the thick capsule of K.
pneumoniae has been shown to impede the activity of type 1 fimbriae and also to
retard the assembly of type 1 fimbrial subunits from periplasm to cell surface (204, 260, 267), suggesting a cross-regulation of the expression of fimbriae and capsule for an efficient infection.
1.2.2. K. pneumoniae type 3 fimbriae
Type 3 fimbriae are 2-4 nm wide and 0.5-2 μm long surface organelles that are originally characterized in Klebsiella strains by their ability to mediate mannose-resistant agglutination of tannic acid-treated human erythrocytes (MR/K haemagglutination) (78, 106). Several studies have also demonstrated an important role for type 3 fimbriae in biofilm formation on biotic and abiotic surfaces (33, 39, 71, 148, 175, 232, 234, 273, 291). Biofilms are recognized as surface-attached bacteria embedded in a self-produced matrix, composed mainly of polysaccharide, but also containing proteins and nucleic acids (292). Biofilm formation promotes encrustation and protects the bacteria from the hydrodynamic forces of urine flow, host defenses
and antibiotics (316). The ability of bacteria to form biofilm on medical devices is believed to play a major role in development of nosocomial infections, including the catheter-associated urinary tract infections, which is frequently caused by K.
pneumoniae (200, 244, 251, 316). In addition, type 3 fimbriae mediate adhesion to
epithelial cells, from the respiratory and urinary tracts, and extracellular matrix proteins, such as collagen V, in vitro (133, 140, 148, 277, 296, 297).
Type 3 fimbriae are encoded by the mrkABCDF operon (8, 78, 139). MrkA and MrkF are the major and minor subunits, respectively, which constitute the fimbrial rod and facilitate biofilm formation (139, 175). MrkD is the adhesin that mediates binding specificity and biofilm formation on extracellular matrix-coated surfaces (134, 139, 148, 275), however its cognate receptor is still unknown. MrkB and MrkC are predicted to be chaperone and usher, respectively, which are responsible for the fimbrial assembly. A putative regulatory gene, mrkE, located upstream to mrkA has also been reported in K. pneumoniae IA565, which harbors a plasmid carrying the type 3 fimbrial genes (8). The mrk genes have been shown to reside at multiple genomic locations, including the chromosome (291), on conjugative plasmids (39, 232) and within a composite transposon (225). The spread of the type 3 fimbrial gene between Enterobacteriaceae strains by lateral gene transfer has also been described (39, 233).
1.3. Cyclic-di-GMP signaling
Bis-(3’-5’)-cyclic dimeric guanosine monophosphate (Cyclic-di-GMP or c-di-GMP), has emerged as a second messenger specific to the domain of Bacteria (151, 239, 250, 256, 293). c-di-GMP controls a variety of cellular processes, mainly biogenesis and function of extracellular components, flagella and fimbriae, and exopolysaccharide synthesis. The intracellular level of c-di-GMP in bacteria is modulated through the activity of di-guanylate cyclases (DGCs) that convert two molecules of GTP to c-di-GMP, and phosphodiesterases (PDEs) that linearize c-di-GMP to pGpG, which is subsequently hydrolyzed to GMP. DGCs are characterized by the active site GG[D/E]EF amino acid motif in the enzyme catalytic site (127, 201, 237, 259), whereas PDEs contain either the EAL domain or HD-GYP domain (45, 57, 255, 271, 294). Formation of c-di-GMP requires dimerization of two GGDEF domains, and c-di-GMP degradation can be mediated either by the EAL domain or by the less common HD-GYP domain. DGCs and PDEs usually harbor an N-terminal signal input domain that regulates the activity of its C-terminal catalytic GGDEF or EAL/HD-GYP domain (122, 151, 269). Besides, GGDEF domains are often found together with EAL or HD-GYP domains in a single polypeptide. The coexistence of opposing enzymatic activities in these ‘hybrid’ proteins has long been controversial and only a few reports suggest that bifunctional proteins may exist (92,
168, 298). It has also been demonstrated that degenerated GGDEF domain may retain substrate-binding capacities and provide GTP-dependent control of the activity of an EAL domain in a single peptide (57, 137, 158). The mechanism of the intramolecular coordination of GGDEF and EAL/HD-GYP domains in composite protein remains to be shown.
GGDEF and EAL domain proteins are ubiquitous in bacteria but absent from archaea (103, 256). A single bacterial genome generally encodes many different members of these protein families (e.g. E. coli harbors 19 GGDEF and 17 EAL genes;
Vibrio cholerae harbors 41 GGDEF, 22 EAL, and 9 HD-GYP genes) (103). Moreover,
genomes were found to encode several GGDEF and EAL domain proteins with a particularly striking expansion in γ-Proteobacteria. This highly redundant and complex system are suggested to achieve signaling specificity through different modes of sequestration, including microcompartmentalization, temporal regulatory sequestration, and co-localization of the DGCs, PDEs, effector, and target molecules that constitute functional c-di-GMP signaling modules (151, 156, 238, 239, 256, 286, 317). A computational analysis of 11248 GGDEF and EAL domain proteins in 867 prokaryotic genomes also suggests that post-translational regulation and catalytic activity of these proteins play important roles in c-di-GMP signaling (278). As shown in Fig 1.1, in the genome of K. pneumoniae NTUH-K2044, ORFs encoding 11
GGDEF, 10 EAL, and 5 GGDEF-EAL domain proteins were found using a HMMER search (24, 81).
The response to fluctuating cellular levels of c-di-GMP is mediated by a variety of specific effector proteins or RNAs that control specific cellular processes (269). Four types of c-di-GMP effector proteins are currently known (122), and the most prevalent example of such effectors are c-di-GMP-binding proteins harboring a PilZ domain (12). The PilZ domain proteins studied so far seem to be activated by c-di-GMP and to function by protein-protein interactions (34, 122). In some cases, the PilZ domain is directly attached to the C-terminus of the GGDEF, EAL and/or HD-GYP domains (12), or is linked to a domain that generates a molecular output (12, 210, 231). BcsA, the catalytic subunit of cellulose synthase from Gluconacetobacter
xylinus, is the first identified PilZ domain protein and activated via binding to
c-di-GMP (205, 253, 319). The interaction of c-di-GMP with PilZ domains is further supported by binding and mutagenesis studies of several PilZ domain proteins (34, 56, 210, 245, 258). These studies demonstrate c-di-GMP binding with sub-micromolar affinity dependent on residues in the RxxxR and D/NxSxxG sequence motifs conserved in PilZ domains (27). The high-affinity binding of c-di-GMP of PilZ domain protein is also supported by NMR studies of PA4608 from Pseudomonas
to regulate flagellar activity (34, 56), twitching motility (9), alginate synthesis (210), biofilm formation, and virulence in different bacteria (245). In K. pneumoniae NTUH-K2044, three PilZ domain proteins were found (Fig. 1.1); two of them are contained in the C-terminus of putative cellulose synthases, and the other one, named MrkH (153), is a putative protein of unknown function.
1.4. Thesis objectives
Fimbriae are the most well-known adherence factors in Gram-negative bacteria and play a crucial role in the initial step of infection, thus we focused on the study of
K. pneumoniae fimbrial adherence in this thesis. The objectives are to investigate the
prevalence and expression of fimbriae in K. pneumoniae for a better understanding of its pathogenic mechanism. Two highly virulent K. penumoniae strains isolated from liver abscess, NTUH-K2044 and CG43 (belonged to K1 and K2 serotypes, respectively), are studied in parallel. The study flow is listed as following:
Chapter 2 reports the identification of putative fimbrial gene clusters in the genome of K. pneumoniae NTUH-K2044. Besides type 1 and type 3 fimbrial genes, the others are novel and were designated Kpa, Kpb, Kpc, Kpd, Kpd, Kpf, and Kpg fimbriae. Prevalence analysis of the nine fimbrial gene clusters among K. pneumoniae clinical isolates was performed by PCR detection.
Chapter 3 characterizes the Kpc fimbriae which is prevalent in K. pneumoniae strains of serotype K1. Expression of Kpc fimbriae was found to increase bacterial biofilm-forming activity. A recombinase-mediated phase variation of the Kpc fimbriae expression is also elucidated.
Chapter 4 presents analyses of the regulation of type 3 fimbriae expression. Roles of three genes encoding putative regulators, MrkH, MrkI, and MrkJ, on the expression of type 3 fimbriae were studied. A global iron uptake regulator, Fur, also appeared to affect the type 3 fimbriae expression, and this regulation was found to be mediated by the c-di-GMP signaling. Furthermore, environmental stimuli including Fe2+ and
oxygenation that influence the expression of type 3 fimbriae were analyzed.
Chapter 5 concludes with a comprehensive view and provides perspectives regarding further investigations on the fimbrial adherence of K. pneumoniae.
(A) KP1_2678 GGDEF KP1_2678 GGDEF KP1_1983 GGDEF KP1_1983 GGDEF KP1_3053 GGDEF KP1_3053 GGDEF KP1_3555 GGDEF KP1_3555 GGDEF KP1_4180 GGDEF HAMP KP1_4180 GGDEF HAMP KP1_0227 GGDEF Cache_1 KP1_0227 GGDEF Cache_1 KP1_2841 GGDEF PAS-4 KP1_2841 GGDEF PAS-4 KP1_0782 GGDEF KP1_0782 GGDEF GGDEF KP1_1864 GGDEF KP1_1864 GGDEF GGDEF KP1_2191 GGDEF GAF KP1_2191 GGDEF GAF GGDEF GAF KP1_3652 GGDEF GAF KP1_3652 GGDEF GAF GGDEF GAF (B) KP1_4554 (MrkJ) EAL KP1_4554 (MrkJ) EAL EAL KP1_4578 (FimK) EAL KP1_4578 (FimK) EAL KP1_1729 EAL BLUF KP1_1729 EAL BLUF EAL BLUF KP1_3839 EAL KP1_3839 EAL KP1_1978 EAL KP1_1978 EAL KP1_0324 (YjcC) EAL KP1_0324 (YjcC) EAL EAL KP1_3456 EAL KP1_3456 EAL EAL KP1_1299 EAL KP1_1299 EAL KP1_4080 EAL MASE1 KP1_4080 EAL MASE1 EAL MASE1 KP1_2624 (BlrP1) EAL BLUF KP1_2624 (BlrP1) EAL BLUF (C) KP1_5220 GGDEF HAMP EAL KP1_5220 GGDEF HAMP EAL KP1_3659 GGDEF EAL KP1_3659 GGDEF EAL KP1_2724 GGDEF PAS EAL KP1_2724 GGDEF PAS EAL KP1_3998 GGDEF MASE1 EAL KP1_3998 GGDEF
MASE1 GGDEF EAL
MASE1 EAL KP1_4976 GGDEF EAL KP1_4976 GGDEF EAL GGDEF EAL (D) KP1_4551 PilZ KP1_4551 PilZ KP1_5237 (AcsA) PilZ
Glycosyl transferase family 2
KP1_5237 (AcsA)
PilZ
Glycosyl transferase family 2
KP1_5225 (BcsA)
PilZ
Glycosyl transferase family 2
KP1_5225 (BcsA)
PilZ
Fig. 1.1. Domain architecture of putative c-di-GMP signaling proteins encoded by the K. pneumoniae NTUH-K2044 genome. The locus tag (KP1_number) of the genes encoding (A) GGDEF, (B) EAL, (C) GGDEF/EAL, or (D) PilZ domain proteins was indicated. Analysis of protein functional domain, as indicated, was performed using the Pfam database provided online (http://www.sanger.ac.uk /Software/Pfam/). The identities of the proteins are also shown in blue: four EAL domain proteins (YjcC, FimK, MrkJ, and BlrP1) that have been described in K.
pneumoniae (22, 153, 170, 252) and two PilZ domain proteins (putative cellulose
synthases BcsA and AcsA) (249). Predicted domain with unknown function is shown by a twilled box.
CHAPTER 2
Prevalence Analysis of the Fimbrial Gene
Clusters in Klebsiella pneumoniae
2.1. Abstract
Using HMMER search for genes encoding the Pfam fimbrial components in the genome of K. pneumoniae NTUH-K2044, nine distinct fimbrial gene clusters were identified. In addition to fim and mrk gene clusters, encoding type 1 and type 3 fimbriae respectively, the other seven are novel and named kpa, kpb, kpc, kpd, kpe, kpf, and kpg. The presence of the fimbrial genes among 105 K. pneumoniae strains isolated from various infection sites was analyzed by PCR detection, and the kpb and
kpc genes were found to be more prevalent (P < 0.0001) in the isolates of serotype K1.
Besides, an RFLP analysis was performed among the 105 K. pneumoniae isolates which revealed most of the isolates possess a v1-like mrkD RFLP type.a
a A part of this chapter has been published:
Wu, C. C., Y. J. Huang, C. P. Fung, and H. L. Peng. 2010. Regulation of the
Klebsiella pneumoniae Kpc fimbriae by the site-specific recombinase KpcI.
2.2. Introduction
A large number of fimbrial gene clusters are commonly present in a bacterial genome (192, 301, 305), which are believed to be expressed differentially in order to adhere to various host receptors during infection (144). In Salmonella enterica serovar Typhi (S. Typhi) CT18, 14 fimbrial gene clusters including a type 4 fimbrial operon, an orthologue of the agf (csg) operon, and 12 putative fimbrial operons of the chaperone-usher assembly class were identified (301). Prevalence study among different Salmonella isolates has revealed that an unique repertoire of fimbrial gene clusters is possessed by the strains of serotype Typhi, which is probably resulted in specific host adaptation (301).
Among these Salmonella fimbriae, only two of them, fim and agf (csg), have been demonstrated to express on the surfaces of serotype Typhimurium cells by electron microscopy (79, 112, 190, 289). The remaining 11 fimbrial operons are poorly expressed when bacteria are grown under standard laboratory conditions (143). However, expression of some of the fimbriae could be detected by flow cytometry while the Typhimurium cells recovered from bovine ligated ileal loops at 8 h after infection (143). Seroconversion to 11 types of fimbriae was found indicating that a transient expression of each of the fimbriae induced the host immune responses (142).
Moreover, the individual deletion of six of the fimbrial operons affected the bacterial persistence in intestines of mice (318), and a strain carrying mutations of four of the fimbrial operons resulted in a 26-fold increase of LD50 to mice (306).
Cross-regulation between these fimbrial operons have also been described (130, 131, 326). Mutation of a fimbrial operon could activate the expression of another type of fimbriae (54, 274), further supporting a regulatory network involved in the expression of the multiple fimbriae.
Sequence variation of the fimbrial adhesin affects not only binding specificity but also fimbrial activity. We have previously found that the mrkD gene, encoding the type 3 fimbrial adhesin, from K. pneumoniae clinical isolates could be classified into four restriction fragment length polymorphism (RFLP) types: mrkDv1, mrkDv2, mrkDv3, and mrkDv4 (140). The MrkD sequence variation determines the binding
specificity and the assembly efficiency of type 3 fimbriae (46, 140).
We initiated the study using bioinformatic tools to identify the fimbrial genes in the genome of K. pneumoniae NTUH-K2044, a liver abscess isolate of serotype K1 (325). Prevalence study of the fimbrial genes was then employed among 105 K.
pneumoniae clinical isolates from different infection sites. Besides, the mrkD-RFLP
2.3. Results
2.3.1. Identification of the fimbrial gene clusters in K. pneumoniae NTUH-K2044
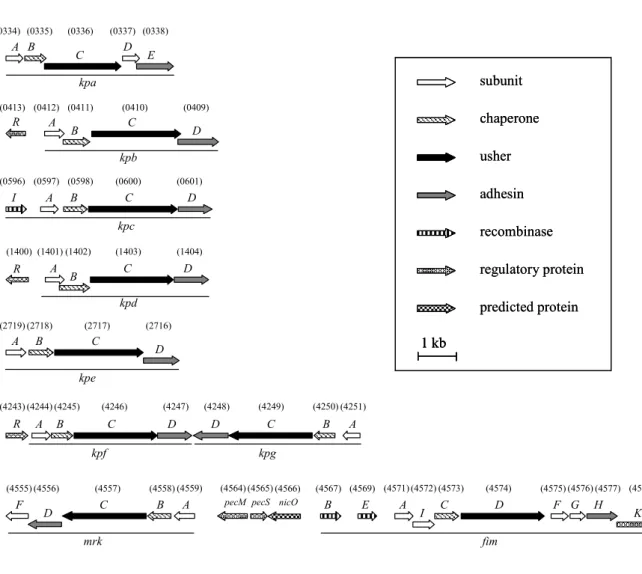
Nine fimbrial gene clusters were identified using the HMMER search of the genome of K. pneumoniae NTUH-K2044. Each contained at least four genes, encoding a putative major pilin, a chaperone, an usher and an adhesin for the biosynthesis of fimbriae belonging to the chaperone-usher assembly class. As shown in Fig. 2.1, these include the type 1 and type 3 fimbrial gene clusters fim and mrk, and seven novel ones, namely kpa, kpb, kpc, kpd, kpe, kpf, and kpg. Multiple sequence alignment by CLUSTAL W showed that the amino acid sequences of these pilins and adhesins shared 26.5-36.4% similarity; chaperones and ushers shared a higher similarity ranging from 49.3 to 55.4%. The mrk-fim fimbrial genes are clustered and transcribed divergently (Fig. 2.1). This gene organization including the pecM, pecS and nicO homologues has been found to be conserved in the genomes of K.
pneumoniae CG43, C3091, MGH78578 and 342 (95, 230, 291). The gene clusters kpf
and kpg are also linked physically but transcribed convergently.
BLAST analysis, using the sequences of the nine fimbrial gene clusters identified in K. pneumoniae NTUH-K2044 as templates, of the genome of K. pneumoniae strains MGH78578 and 342 (95, 230) showed that, except for kpc and kpf, the genes
were conserved in the three genomes. No homologue of the kpc genes was found in the strains MGH78578 and 342, while homologues of the kpf genes were found in the genome of MGH78578 but not of 342. Besides, except for the kpc genes, the other eight fimbrial gene clusters were also identified in the contig sequences of CG43 (unpublished results from Dr. S.-F. Tsai, National Health Research Institutes, Taiwan).
2.3.2. PCR screening for the presence of the fimbrial genes
To investigate the prevalence of the nine fimbrial gene clusters among K.
pneumoniae strains, a total of 105 K. pneumoniae clinical isolates, namely the TVH
strains, from different infection sites were collected. Two specific primer pairs corresponding to the pilin- and adhesin-encoding genes were designed for PCR detection. Prevalence was determined on the basis of the presence of the PCR amplicons (Fig. 2.2). The analysis revealed the presence of kpa, kpd, kpe, kpg, fim, and mrk genes in most of the isolates, and the prevalence percentages were 99, 82, 93, 97, 84 and 100%, respectively. The prevalence percentages for the kpb, kpc and kpf genes were lower at 52, 33 and 70% of the isolates, respectively. No obvious correlation between fimbrial type and disease could be identified. However, the kpb and kpc genes were shown to be more prevalent in K1 isolates (P<0.0001). As shown in Table 2.1, most of the clinical isolates of serotype K1 harbored kpb and kpc genes,
while non-K1 isolates carrying the kpb and kpc genes were much less frequent (32 and 1%, respectively). The close association of Kpc fimbriae with serotype K1 prompted the selection of the Kpc fimbriae for further study of the K. pneumoniae liver abscess pathogenic mechanism (Chapter 3).
2.3.3. PCR-RFLP analysis of the mrkD genes
A PCR-RFLP analysis previously described (140) was also employed to determine the mrkD type of the K. pneumoniae clinical isolates. The result showed that 87 of the 105 isolates possess mrkD gene of v1-like RFLP. The number of isolates harbored the v2-, v3-, and v4-like mrkD were respectively four, two, and four, while eight carried a novel mrkD RFLP type. The mrkD-RFLP types could not be associated with certain diseases. Interestingly, sequence variation was found in mrkD genes of same RFLP type. For example, K. pneumoniae NTUH-K2044, CG43, and TVH2 harbor mrkD gene of v1-RFLP type while frameshift mutations were found in mrkD gene of CG43 and TVH2. These sequence variations which resulted in a premature termination of the mrkD translation may affect the type 3 fimbrial assembly and adherence properties as reported (46, 140).
2.4. Discussion
Unlike some of the fimbrial gene clusters in S. Typhi CT18 and in Escherichia
coli O157:H7 that contain either premature termination codons or frameshift
mutations, the nine fimbrial gene clusters identified in K. pneumoniae NTUH-K2044 appeared to be intact. Although these fimbrial gene clusters identified using bioinformatic tools are putative ones, we anticipate that, besides the type 1 and type 3 fimbriae, the other seven are required for K. pneumoniae NTUH-K2044 infection at not yet identified environments.
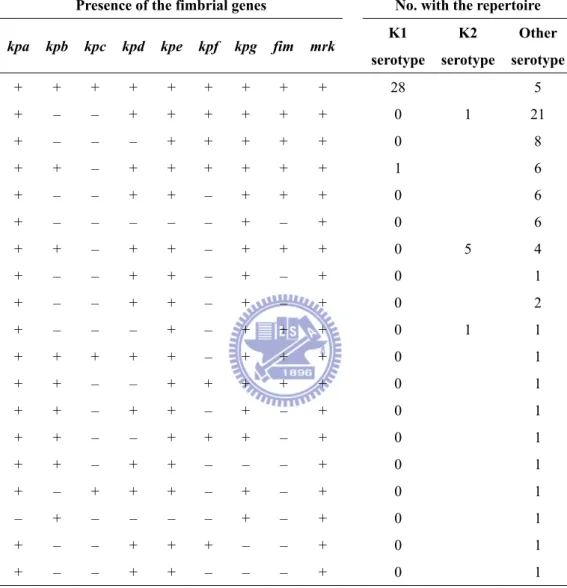
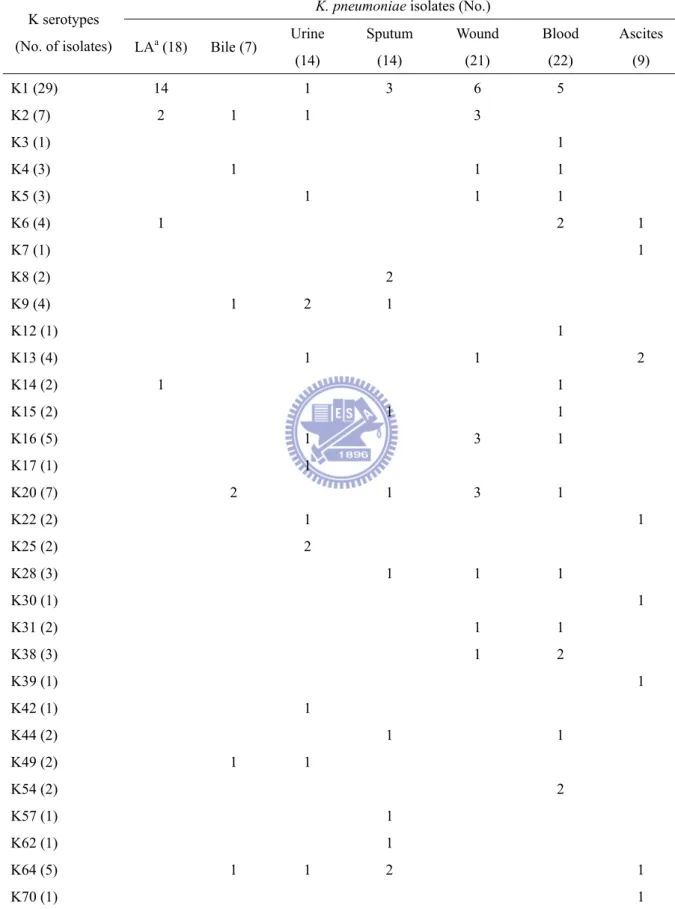
A specific repertoire of fimbrial operons has been proposed as a complex virulence factor involved in S. Typhi infections (144). Although no obvious correlation between the disease and fimbrial type was noted, 28 out of 29 K1 isolates possessed an identical repertoire of fimbrial gene clusters suggesting a role of the fimbrial repertoire in pathogenicity of K1 isolates (Table 2.2). Consistent with the other reports (100, 178), most of the liver abscess isolates (14 out of 18) were capsular serotype K1 (Table 2.3). Aside from that, the capsular serotypes of the K.
pneumoniae isolates appeared no correlation with the infection sites (Table 2.3).
As shown in Fig. 2.1, three ORFs encoding putative transcriptional factors, located upstream to the kpb, kpd, and kpf fimbrial genes, were designated kpbR, kpdR,
and kpfR, respectively. Regulator encoding genes are commonly found to be located adjacent to fimbrial operons (17, 94, 125, 141), such as papB for the E. coli P fimbrial gene cluster. PapB not only regulates the expression of P fimbriae but also affects the type 1 fimbriae expression (130, 131, 326). Besides, a mutant with a Tn5 insertion in the putative promoter region of kpgA decreased the expression of type 3 fimbriae in K.
pneumoniae 43816 (33). These findings suggested a cross-talk regulation is generally
present for the control of the expression of different fimbriae. Nevertheless, functional roles of these putative fimbriae and their cognate regulators remain to be studied.
In many cases, mutation of a fimbrial operon did not affect or only moderately altered the bacterial virulence (25, 26, 191, 318). It may be due to a suboptimal experimental model or the gene-loss was functionally compensated by other adherence factors. It is also believed that multiple fimbriae may act synergistically or differentially to adhere to host cells during infection. Besides S. Typhi and Typhimurium, to our knowledge, genomic analysis of fimbrial gene clusters has also been described in Pseudomonas aeruginosa and E. coli O157:H7 (192, 222, 305). Herein, the genome-wide analysis of fimbrial gene clusters paved the way for further studies of K. pneumoniae fimbrial adherence.
Table 2.1. Prevalence of kpb and kpc genes in K. pneumoniae isolates
No. of kpb strain/total no. of strain isolated (%) No. of kpc strain/total no. of strain isolated (%)
K. pneumoniae
isolate K1 serotype other serotype K1 serotype other serotype
LAa 14/14 3/4 14/14 1/4 Bile 2/7 1/7 Urine 1/1 3/13 1/1 0/13 Sputum 3/3 3/11 2/3 1/11 Wound 6/6 5/15 6/6 1/15 Blood 5/5 4/17 5/5 2/17 Ascites 4/9 1/9 Total 29/29 (100)* 24/76 (32)* 28/29 (97)* 7/76 (1)* *, P < 0.0001; a, Liver abscess
Table 2.2. Repertoire of fimbrial genes among K. pneumoniae isolates with different K serotypes
Presence of the fimbrial genes No. with the repertoire
kpa kpb kpc kpd kpe kpf kpg fim mrk K1
serotype K2 serotype Other serotype + + + + + + + + + 28 5 + – – + + + + + + 0 1 21 + – – – + + + + + 0 8 + + – + + + + + + 1 6 + – – + + – + + + 0 6 + – – – – – + – + 0 6 + + – + + – + + + 0 5 4 + – – + + – + – + 0 1 + – – + + – + – + 0 2 + – – – + – + + + 0 1 1 + + + + + – + + + 0 1 + + – – + + + + + 0 1 + + – + + – + – + 0 1 + + – – + + + – + 0 1 + + – + + – – – + 0 1 + – + + + – + – + 0 1 – + – – – – + – + 0 1 + – – + + + – – + 0 1 + – – + + – – – + 0 1
Table 2.3. K serotypes of the K. pneumoniae clinical isolates
K. pneumoniae isolates (No.)
K serotypes
(No. of isolates) LAa (18) Bile (7) Urine
(14) Sputum (14) Wound (21) Blood (22) Ascites (9) K1 (29) 14 1 3 6 5 K2 (7) 2 1 1 3 K3 (1) 1 K4 (3) 1 1 1 K5 (3) 1 1 1 K6 (4) 1 2 1 K7 (1) 1 K8 (2) 2 K9 (4) 1 2 1 K12 (1) 1 K13 (4) 1 1 2 K14 (2) 1 1 K15 (2) 1 1 K16 (5) 1 3 1 K17 (1) 1 K20 (7) 2 1 3 1 K22 (2) 1 1 K25 (2) 2 K28 (3) 1 1 1 K30 (1) 1 K31 (2) 1 1 K38 (3) 1 2 K39 (1) 1 K42 (1) 1 K44 (2) 1 1 K49 (2) 1 1 K54 (2) 2 K57 (1) 1 K62 (1) 1 K64 (5) 1 1 2 1 K70 (1) 1 a Liver abscess
subunit chaperone usher adhesin recombinase regulatory protein predicted protein 1 kb subunit chaperone usher adhesin recombinase regulatory protein predicted protein 1 kb 1 kb kpa A B C D E kpb A B C D kpd A B C D kpc A B C D I A B C kpe D kpf kpg A B C D D C B A mrk fim F C B A D pecM pecS B E A I C D F G H K (0334) (0335) (0336) (0337) (0338) (0413) (0412) (0411) (0410) (0409) (059 (0597) (0598) (0600) (0601) (1 ) (1401) (1402) (1403) (1404) (271 (2718) (2717) (2716) (424 (4244) (4245) (4246) (4247) (4248) (4249) (4250) (4251) (4 ) (4556) (4557) (4558) (4559) (4564) (4565) (4567) (4569) (4571)(4572) (4573) (4574) (4575) (4576) (4577) (4578) R 6) 400 R 9) 3) R 555 (4566) nicO
Fig. 2.1. Fimbrial gene clusters of the chaperone-usher-dependent assembly class in K. pneumoniae NTUH-K2044. The designation of putative fimbrial genes and the locus tag (KP1_number) of ORFs annotated in the K. pneumoniae NTUH-K2044 genome are indicated. A total number of nine fimbrial gene clusters and genes encoding putative regulators are as shown. Each of the putative fimbrial operons is underlined. The putative functions of the ORFs are also shown.
1000 750 500 1500 M 1 2 3 4 5 6 7 8 9
*
*
*
*
*
*
*
*
*
Δ Δ Δ Δ Δ Δ Δ Δ Δ bp 1000 750 500 1500 M 1 2 3 4 5 6 7 8 9*
*
*
*
*
*
*
*
*
Δ Δ Δ Δ Δ Δ Δ Δ Δ bpFig. 2.2. PCR amplicons of the pilin and adhesin encoding genes in K. pneumoniae NTUH-K2044. Lane M: Gene Ruler 1-kb DNA molecular size markers
(Fermentas, Vilnius, Lithuania). The DNA fragments represent pilin (Δ) and adhesin (*) encoding genes, lanes 1 to 9, respectively of Kpa, Kpb, Kpc, Kpd, Kpe, Kpf, Kpg, type 3, or type 1 fimbriae.
CHAPTER 3
Regulation of Kpc Fimbriae by the Site-specific
Recombinase KpcI
3.1. Abstract
In this study, the Kpc fimbria of Klebsiella pneumoniae NTUH-K2044 was characterized. Induced expression of the recombinant kpcABCD genes in Escherichia
coli resulted in Kpc fimbriation and increased biofilm formation, suggesting that the kpc genes are sufficient to encode a functional fimbrial apparatus. A putative
site-specific recombinase encoding gene kpcI and a 302-bp intergenic DNA flanked by 11-bp inverted repeats, namely kpcS, were identified in the upstream region of the
kpcABCD genes. Using LacZ as the reporter, a dramatic difference in promoter
activity of kpcS in two different orientations was observed and assigned as ON and OFF phase accordingly. Expression of kpcI appeared to be able to invert the kpcS in
trans from phase OFF to ON and vice versa. Using the two plasmid system,
expression of kpcA, encoding the major component of the Kpc fimbriae, could be observed upon the induced expression of kpcI. These results indicate that KpcI is involved in the regulation of Kpc fimbriation in a phase-variable manner.a
a A part of this chapter has been published:
Wu, C. C., Y. J. Huang, C. P. Fung, and H. L. Peng. 2010. Regulation of the
Klebsiella pneumoniae Kpc fimbriae by the site-specific recombinase KpcI.
3.2. Introduction
Expression of Escherichia coli type 1 and P fimbriae are regulated by a mechanism called phase variation, which referred to a reversible switch between “all-or-none” (ON/OFF) expressing phase (116, 121, 307). The phase variation, resulted in variation of fimbriae expression between individual cells of a clonal population, is a genetic or epigenetic mechanism that allows the variability to be heritable. Phase-expression is reversible between generations, and the reversion frequency exceeds that of a random mutation (116, 121, 307).
The phase-variation control of different fimbriae include conservative site-specific recombination (CSSR) for type 1 and MR/P fimbriae (4, 29, 181, 334), DNA methylation for P and Pef fimbriae (36, 125, 126, 223), and slipped-strand mispairing (SSM) for fimbriae of Bordetalla pertusis and Haemophilus influenzae (308, 320). The E. coli type 1 fimbriae represent the best-studied module for CSSR-mediated fimbriae expression. The oscillating ON-and-OFF expression of type 1 fimbriae is correlated with the inversion of a 314-bp DNA sequence (fimS) immediately upstream of fimA, the major pilin encoding gene. The promoter located within fimS that drives the expression of the type 1 fimbriae is flanked by 9-bp inverted repeats (IRs). The fim operon is only expressed when the fimS is in the ON
orientation, which allows transcription of the fim operon. The two genes located upstream to fimS respectively encode FimB and FimE site-specific DNA recombinases of the λ integrase family (86, 227). The recombinase FimB inverts fimS in the ON-to-OFF and OFF-to-ON directions, whereas FimE determines predominantly the ON-to-OFF direction of fimS (31, 101, 206). As a result, alternations of the expression ratio of FimB and FimE modulate the phase variation of type 1 fimbriae (1, 75, 283, 321). Other types of fimbriae controlled by DNA recombinase-mediated phase variation include Proteus mirabilus MR/P fimbriae (181, 334), enterotoxigenic E. coli CS18 fimbriae (132), and Photorhabdus Mad fimbriae (287). Unlike type 1 fimbriae, the phase variation of MR/P and Mad fimbriae is controlled by only one site-specific recombinase, MrpI and MadR, respectively, which catalyzed the DNA inversion in both directions (181, 287, 334).
Prevalence study among K. pneumoniae isolates has demonstrated that the putative fimbrial genes kpcABCD are highly prevalent in isolates of capsular serotype K1 (Table 2.1). In addition, a gene encoding a putative DNA recombinase was identified in the upstream region of the kpcABCD genes (Fig. 2.1) and was named
kpcI. Whether KpcI modulates a phase variable expression of kpcABCD genes is
3.3. Results
3.3.1. Display of the Kpc fimbriae on E. coli surface
In order to determine whether the kpcABCD genes encode a fimbrial apparatus, an E. coli fimbriae display system was used. The kpcABCD genes were PCR-amplified and cloned into pET30a, designated pKPC-7, to allow controlled expression by IPTG induction. Numerous thin and rigid fimbriae on the surface of E.
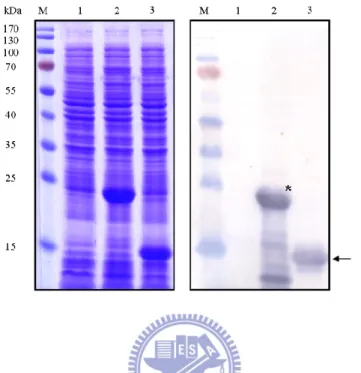
coli Novablue (DE3) harboring pKPC-7 could be observed, while the E. coli
harboring pET30a was afimbriate (Fig. 3.1). Expression of KpcA, the putative major pilin, could also be detected by Western blot analysis (Fig. 3.2). These results indicated that the kpcABCD genes are sufficient to produce a fimbrial apparatus. However, the growth rate of E. coli harboring pKPC-7, in comparison with that of E.
coli Novablue (DE3) [pET30a] was obviously decreased even without IPTG induction.
This could be a result of the overexpression of kpc genes, especially the kpcA (Fig. 3.2, lane 3). In case the biased growth rate impeded the characterization of the Kpc fimbriae, another expression plasmid pETQ that can be used for protein expression under T5lac promoter control in E. coli and K. pneumoniae was used. The pETQ plasmid carrying the kpcABCD genes, pKPC-36, was then transformed into E. coli HB101 which is an afimbriate bacterium. Expression of the kpcABCD genes was
tightly controlled under the T5lac promoter, and no obvious difference in growth rate between E. coli HB101 harboring pKPC-36 and E. coli HB101 harboring pETQ was noted. Using polyclonal KpcA antiserum, Western blot analysis revealed that KpcA was expressed (Fig. 3.3A), and fimbriation on the surface of the recombinant E. coli HB101 was also observed by fluorescence microscopy (Fig. 3.3B). E. coli HB101 [pKPC-36] was thus used for the characterization of the recombinant Kpc fimbriae.
3.3.2. Expression of Kpc fimbriae increased biofilm-forming activity
Activity assessment of the recombinant Kpc fimbriae, including hemagglutination, cell adherence, and biofilm formation, was subsequently carried out. However, no hemagglutination activity against red blood cells from guinea pig, rabbit, or human (type A and type B) could be observed for the recombinant bacteria. Besides human epithelial cell lines Int407 (intestine), HCT-8 (intestine), Hep-2 (larynx), and T24 (bladder), two human hepatocellular liver carcinoma cell lines, HepG2 and SK-HEP-1, were also used to assess the cell adherence activity of the Kpc fimbriae. The cell adherence assay was performed as described by Huang et al. (140). Although the recombinant E. coli HB101 exhibited differential adherence to these cell lines, Kpc fimbriation did not increase the adherence activity of the bacteria to any of the cells. On the other hand, as shown in Fig. 3.4, E. coli HB101 harboring pKPC-36