行政院國家科學委員會專題研究計畫 成果報告
生物文獻知識庫之建構
計畫類別: 個別型計畫 計畫編號: NSC91-2213-E-009-082- 執行期間: 91 年 08 月 01 日至 92 年 07 月 31 日 執行單位: 國立交通大學資訊科學學系 計畫主持人: 梁婷 計畫參與人員: 王建邦,趙志乾,游志銘,陳建行 報告類型: 精簡報告 處理方式: 本計畫涉及專利或其他智慧財產權,2 年後可公開查詢中 華 民 國 92 年 10 月 27 日
I
行政院國家科學委員會補助專題研究計畫
5 成 果 報 告
□期中進度報告
生物文獻知識庫之建構
Knowledge Base Construction from Biological Texts
計畫類別:
5
個別型計畫 □ 整合型計畫
計畫編號:NSC - 91 - 2213 - E - 009 - 082
執行期間: 91 年 8 月 1 日至 92 年 7 月 31 日
計畫主持人:梁婷
共同主持人:
計畫參與人員:王建邦,趙志乾,游志銘,陳建行
成果報告類型(依經費核定清單規定繳交):5精簡報告 □完整報告
本成果報告包括以下應繳交之附件:
□赴國外出差或研習心得報告一份
□赴大陸地區出差或研習心得報告一份
□出席國際學術會議心得報告及發表之論文各一份
□國際合作研究計畫國外研究報告書一份
處理方式:除產學合作研究計畫、提升產業技術及人才培育研究計畫、
列管計畫及下列情形者外,得立即公開查詢
□涉及專利或其他智慧財產權,□一年
5
二年後可公開查詢
執行單位:交通大學資訊科學系
中 華 民 國 92 年 10 月 27 日
II 一、中文摘要 隨著線上文獻快速成長,如何將文獻中相關資訊自動萃取出,以成為加值型的知識庫 是目前資訊萃取技術一項重要的研究課題。另一方面生物科技產業亦成為新世紀最重要的 工業發展項目。有鑑於此,在本計劃中我們將針對大量生物文獻提出一個線上知識庫建構 系統以確實有效地將文獻資料中的資訊轉換成結構化的資料型式以便於資訊擷取。 知識庫建構系統將包含三個主要模組分別是檢索詞庫建構模組、資訊擷取模組、和資 訊萃取模組。檢索詞庫建構模組處理生物體名辨識和收集、相關之關鍵詞萃取、及詞群建 立。資訊擷取模組包含查詢自動擴充及資訊過濾處理;資訊萃取模組則主要將文句中所敘 述的生物資訊萃取出來。 在本計畫中我們利用統計方法自動建立三個索引詞庫包括 MeSH 詞群庫、細菌類別關鍵 詞庫、和以主要動詞為主的句型庫。實驗證明這三個新的知識庫有助於文獻的自動檢索、 分類和查詢。另方面所提的單一介面的查詢系統可簡化查詢者對近端和遠端資料庫的查詢 工作。此系統並嵌入一個查詢相關判讀機制及文獻線上資訊萃取器以便於知識的管理。所 提的方法亦可適用於其他生物領域。 我們希望藉由此計劃的執行,一方面開發出有效可行的資訊萃取法則將大量的生物文 獻資料轉換成加值型的知識庫;此外,亦提供生物學家一個有效的知識萃取與處理系統, 以促進生物資訊的探勘。 關鍵詞:資訊萃取、資訊擷取,檢索詞庫,知識庫,生物文獻 二、英文摘要
As more and more scientific papers become available on-line, there is a growing need for systems that can transform textual data into value-added knowledge bases. On the other hand the development of biological technologies has become one of the most important issues in this new century. In view of this an empirical system that can inexpensively and accurately map the information of biological texts into a structured representation will be studied and designed in this proposal.
The proposed on-line knowledge base construction system will contain three major components, namely, the thesaurus construction module, the information retrieval module and the information extraction module. The thesaurus construction module involves material name identification and collection, associated key terms prediction and term cluster generation. The information retrieval module concerns the automatic query expansion and information filtering. The information extraction module addresses the identification of the domain-specific relation between biological objects.
In this project three new thesauri, namely bacteria-related MeSH clusters, bacteria descriptors and verb-based informative patterns, are constructed on the basis of statistical approaches. Experimental results with real corpus prove that they are very useful at document
III
indexing, categorization and retrieval. On the other hand a unified web-based retrieval subsystem is implemented to ease user’s request task at dealing with databases either local sites or remote sits. The subsystem is also embedded with a ranking mechanism to enhance retrieval quality and an on-line information extractor to mine useful from document, so as to facilitate knowledge management. The proposed methods are also applicable in other biomedical domains.
Throughout the implementation of this project, novel and empirical extraction methods for large biological information can be explored. In addition, this task will support biologists to ease their knowledge extraction and management, thus facilitating biological research.
Keywords: information extraction, information retrieval, thesaurus construction, text data, knowledge base.
1
二、前言與研究目的
In recent years, the rapid development of computer technologies has facilitated the proliferation of bioinformatics, hence producing lot of research literature. However information in text form, such as MEDLINE records, remains a greatly underutilized source of biological information. Therefore there is growing need for systems that can speedup knowledge acquisition as well as knowledge management for biologists [1, 2, 3, 5, 6, 7, 8, 10, 12]. Hence this project is aimed to develop efficient information retrieval methods and empirical information extraction methods that can inexpensively and accurately construct knowledge base at the end.
Essentially a complete knowledge base construction system will contain three parts, namely, the information retrieval (IR) module, the thesaurus construction (TC) module, and the information extraction (IE) module. The IR module will retrieve the relevant papers of interest from textual databases effectively and exhaustively and the retrieved papers will be categorized into predefined domains and transformed into the corpus for consequent information extraction tasks. The TC module will collect, from the existing databases, the biological terms as well as their associated keywords and it will be updated with the insertion of new lexicons and their related keywords. The IE module will identify the sentences containing biological objects of interest and their relation instances from retrieved paper and transform them into a structured representation of knowledge.
In this project the IR and TC modules will be realized in the first year and the IE module will be implemented in the following year. It is believed that the implementation of this project will be benefit for the tasks such as knowledge database construction and management, summarization and potential scientific discovery.
三、結果與討論
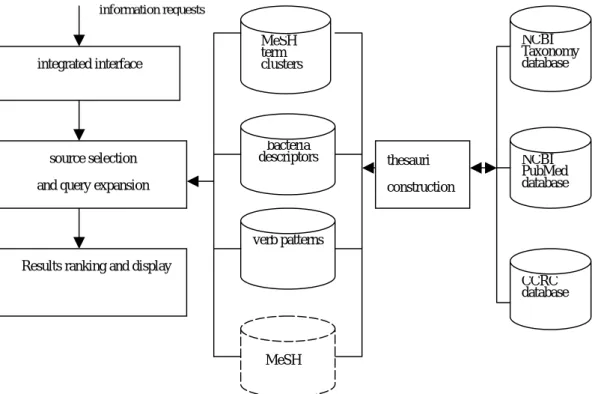
In this project the system is implemented for bacterial domain with the aim to ease bacterial thesaurus construction, information access and management as well. Figure 1 is the system’s overall architecture. It is incorporated with an automatic thesaurus construction module as well as a unified retrieval module. The thesaurus construction is based on statistical methods by using large-scale bacteria corpora which are automatically collected from MEDLINE [8, 10] through PubMed search engine [8]. Three new thesauri are generated, namely, MeSH term clusters, significant bacteria descriptors and bacteria-related verb patterns. Our experimental results showed that the newly created thesauri indeed facilitate document indexing, document categorization, retrieval performance. For example Figure 2 is the similarity distribution between PubMed-indexing set and our indexing set produced by latent semantic analysis. It shows that the proposed bacteria-related MeSH term clusters are useful for document content detection at indexing.
The bacteria descriptor thesaurus is constructed to find the significant terms related to each bacterium so that a new document in bacteria domain can be categorized into and tagged with
2
appropriate bacterial name/class. The performance of the bacteria descriptor finding is verified with a real test data set which are the 3206 articles listed in the reference in Taxonomy. From the experiments, 80% of references are categorized correctly by the system at the first try. 88% references are categorized correctly when their bacteria names appear in the top-five list. Other methods like chi-square test and likelihood ratio test are also implemented for method comparisons. Table 1 shows that the proposed weight-based scheme outperforms the other two methods.
The verb-based pattern extraction from bacteria-related corpus is implemented with the aim to support sentence-like retrieval since there are some verbs playing important roles during information requests in molecular domains. In addition, such verb-based patterns can be viewed as information extraction task which transforms the unstructured textual strings into structured templates. By using the templates the correct answers rather than relevant texts with respect to information requests can be easily accessed and retrieved to enhance retrieval system capability.
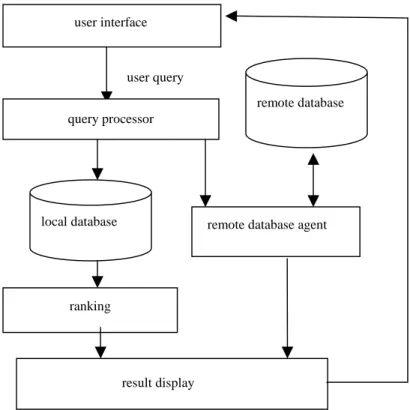
On the other hand the proposed unified retrieval module as shown in Figure 3 simplifies users’ access task to deal with various kinds of databases either at local sites or remote sites. Unlike PubMed which supports sorting-by-attribute display but is lack of ranking functions on retrieved documents, the proposed retrieval module is embedded with a ranking scheme and allows users to browse their requested information detail at their disposal. In addition, any retrieved paper can be processed by the on-line document analyzer such as indexer, bacteria predictor and the pattern extractor, so that its important information can be extracted and be represented with a structured database record. Figure 4 is the result for such processing. All these proposed methods can be easily adaptable to other domains.
四、成果自評
It is believed that the implementation of such system will benefit both information scientist in the context of knowledge discovery and at the same time provide an efficient biological data management and query resolution tools for researchers in microbial strain researchers community. Throughout the implementation of this project four master theses [13-16] and corresponding papers [17, 18] were completed. Other relevant papers based on some part of the project results such as homology search and named entities identification, are still under construction. The support from National Science Council is greatly appreciated.
五、參考文獻
[1] A. R. Aroson, O. Bodenreider, H. F. Chang, S. M. Humphrey, J. G. Mork, S. J. Nelson, T. C. Rindflesch, W. J. Wilbur (2000) “The NLM Indexing Initiative,” AMIA Annual Fall
Symposium, pp.17-21.
[2] J. M. Abasolo and M. Gomez, (2000) “MELISA. An ontology-based agent for information retrieval in medicine,” Proceedings of the First International Workshop on the Semantic Web,
3
pp. 73-82.
[3] P. G.. Baker, C. A. Goble, S. Bechhofer, N. W. Paton, R. Steven and A.Brass(1998) “TAMBIS-Transparent Access to Multiple Bioinformatics Information Sources,” In Proc. of
the 6th International Conference on Intelligent System for Molecular Biology, AAAI Press,
pp. 25-34.
[4] C. C. Chang (1997) “The Study of Chinese Textual Document Retrieval Based on Fuzzy Concept Networks,” Master thesis, Nation Chiao-Tung University.
[5] H. Chen, T. Yim, D.Fye and B.Schatz (1995) “Automatic Thesaurus Generation for an Electronix Community System,” Journal of The American Society for Information Science, Vol. 46, No. 3, pp. 175-193.
[6] B. A. Eckman, A. S. Kosky and L. A. Laroco (2001) “Extending traditional query-based integration approaches for functional characterization of post-genomic data,” Bioinformatics, Vol. 17, No. 7, pp. 587-601.
[7] T. G. Kolda and D. P. O'Leary (1996) “A Semidiscrete Matrix Decomposition for Latent Semantic Indexing in Information Retrieval,” ACM Transactions on Information Systems, Vol. 16, No. 4 pp.322-346.
[8] National Center for Biotechnology Information, (1999) “Entrez”
http://www.ncbi.nlm.nih.gov/Entrez/.
[9] M. F. Porter, (1980) “An algorithm for suffix stripping,” Program, Vol. 14, No. 3, pp. 130-137.
[10] D. L. Wheeler, C. Chappey, A. E. Lash, D. D. Leipe, T. L. Madden, G. D. Schuler, T. A. Tatusova and R. A. Rapp, (2000) “Database resources of the National Center for Biotechnology Information,” Nucleic Acids Res., Vol. 28, pp. 10-14.
[11] J. Xu and W. B. Croft, (1996) “Query expansion using local and global document analysis,”
In Proc. of the ACM-SIGIR Conference on Research and Development in Information Retrieval, Zurich, Switzerland, pp. 4-11.
[12] B. Yates and R. Neto, (1999) “Modern Information Retrieval,” Addison Wesley.
[13] Jia-Kan Chang, (2003) “A Suffix Tree Approach for Homology Search”, Master Thesis, Institute of Computer and Information Science National Chiao Tung University.
[14] Yu-Teng Chang, (2003) “Study of Applying Information Retrieval Techniques to Bacteria Corpus”, Master Thesis, Institute of Computer and Information Science National Chiao Tung University.
[15] Jien-Hsin Chen, (2003) “Named Entity Extraction in Biomedical Domain”, Master Thesis, Institute of Computer and Information Science National Chiao Tung University.
[16] Chih-Ming Yu, (2003) “Relation Extraction from Biological Literature”, Master Thesis, Institute of Computer and Information Science National Chiao Tung University.
[17] Tyne Liang and Yu-Teng Chang, (2003) “A Bacterial Textual Processing and Retrieval System,” accepted by National Computer Symposium.
4
[18] Tyne Liang and Dian-Song Wu, (2003) “Automatic Pronominal Anaphora Resolution in English Texts,” Proceedings of ROCLING XV, pp. 111-127.
5 0 10000 20000 30000 40000 50000 1 0.9 0.8 0.7 0.6 0.5 0.4 0.3 0.2 0.1 0 similarity num be r of a rt ic le s
Figure 2:Similarity between PubMed-indexing set and our-indexing set.
Table 1: The bacterial name tagging accuracy comparison.
rank 1 2 3 4 5
proposed scheme 80% 82% 84% 85% 88%
Chi-square test 46% 72% 78% 80% 82% Likelihood ratio test 25% 50% 58% 71% 75%
thesauri construction MeSH term clusters bacteria descriptors verb patterns MeSH integrated interface NCBI Taxonomy database NCBI PubMed database CCRC database source selection
and query expansion
Results ranking and display
information requests
6
Figure 4: The indexing, prediction and pattern extraction page.
user interface
user query
remote database agent remote database
result display ranking
query processor
local database
7
可供推廣之研發成果資料表
□ 可申請專利 v 可技術移轉 日期:92 年 10 月 27 日國科會補助計畫
計畫名稱:生物文獻知識庫之建構 計畫主持人:梁婷 計畫編號:NSC 91-2213-E-009-082 學門領域:資訊工程技術/創作名稱
Bacterial Document Classification發明人/創作人
Tyne Liang 中文: 以統計方法為主的菌種文獻內容分類法則。此分類法則可在第一次 探測時將 80% 的文獻歸類正確,有助於文獻自動檢索、分類和查詢。技術說明
英文:A statistical bacterial document classifier is designed. 80% of
documents are categorized correctly by the classifier at the first try. It is useful for indexing, classification and retrieval.
可利用之產業
及
可開發之產品
On-line document classification and thesaurus construction.
技術特點
Both algorithms are portable and scalable in other biomedical domains.
推廣及運用的價值
These techniques are the kernel part of the knowledge management related industries.