基因體醫學國家型科技計畫
National Research Program for Genomic Medicine National Science Council, the Executive Yuan, ROC.
探討人類ARL1在運送溶脢體蛋白的功能與機制
Functional characterization of human ARL1 involved in intracellular trafficking: implication in lysosomal disorder
報告類別:□ 新進研究計畫 □ 修正後計畫書 ; 年度成果報告
(New Proposal) (Revised Proposal) (Progress Report)
計畫類別:; 個別型計畫 □ 整合型計畫
(Individual Project) (Program Project)
計畫編號:91GMP032-2
計畫主持人 (Principle Investigator):李芳仁 Fang-Jen S. Lee 共同主持人 (Co-Principle Investigator):
執行單位 (Institution):台大醫學院分子醫學研究所
Institute of Molecular Medicine, College of Medicine, NTU
1
National Research Program for Genomic Medicine National Science Council, the Executive Yuan, ROC.
Progress Report—Research Project 基因體醫學國家型科技計畫 國科會延續性計畫進度報告 個別型計畫 (Individual Project) Program Classification:
; Genomic Medicine Bioinformatics Proteomics & Structural Genomics ELSI
Project Number: 91GMP032-2 (計畫編號) NSC Funding Number: NSC93-3112-B-002-011 (93 年度國科會預核編號) (in Chinese) 中文 探討人類ARL1 在運送溶脢體蛋白的功能與機制 Title of Project 計畫名稱 (in English) 英文
Functional characterization of human ARL1 involved in intracellular trafficking: implication in lysosomal disorder
(in Chinese) 中文
台大醫學院分子醫學研究所 Institution
研究(執行)單位
(in English) 英文
Institute of Molecular Medicine, College of Medicine, NTU (in Chinese) 中文 李芳仁 Principle Investigator 計畫主持人 (in English) 英文 Fang-Jen S. Lee FY 2002 2003 2004 Total Budget 1530000 2080500 2128500 5739000
(in NT dollars: 1USD = 34 NTD) Signature of the PI:_______________Date:_________________
2 TABLE OF CONTENTS
Progress Report ... 3 1. Response to previous reviewers’ critiques... 3
2. Specific Aims... 4
3. Progress Summary ... 6
4. Projected Timeline & Brief Summary of Plans for Next Year ... 31
5. Personnel ... 33
6. Publications and/or Patents... 34
6a. Publications... 34
6b. Patents ... 35 Appendix ... 36
3 Progress Report
1. Response to previous reviewers’ critiques
Please describe the previous reviewers’ critiques and how based on the critiques, you made modifications to specific aims, experimental design, or resource allocation etc.
4 2. Specific Aims
The broad, long-term objective of this proposal is to explore how small GTPases, ADP-ribosylation factor-like proteins (ARL1), with their regulatory proteins, carry out their functions in intracellular Golgi-lysosome trafficking.
The basic membrane recognition and fusion machinery is regulated to fit the needs of individual trafficking steps, specific differentiated cells, and particular physiological states. ADP-ribosylation factors (ARFs) represent a subfamily of Ras-homologous GTPases, including both ARFs and ARF-like proteins (ARLs). ARFs are believed to participate in vesicular transport and membrane trafficking. Recently, we found that human ARL1, similar to yeast ARL1, bound to Golgi and lysosome membranes, consistent with a function in a novel Golgi to lysosome delivery pathway. We hypothesize that ARL1 may interact with their regulatory proteins, participate in membrane trafficking regulation, and thereby may play important roles in lysosomal disorder.
Our specific aims are:
[1] To identify and characterize proteins interacting with or functioning in conjunction with ARL1 in Golgi to lysosome trafficking.
We will use high-affinity double-tags (two IgG-binding units of protein A and calmodulin-binding peptide) fusion protein to purify hARL1-complexes. Functional characterization of these proteins will also give us clues what mechanisms involved in the regulation of intracellular lysosome trafficking. The unidentified abundant specific protein bands will be subjected to LC/MS/MS sequence analysis in the Core laboratory. We anticipate that yeast two-hybrid system will lead to successful identification of genes, which code for proteins interacting with ARL1. We used ARL1 (Q71L) mutant as bait and pulled out five potential ARL1 interacting molecules. We will further use ARL1 (T31N) mutant as a bait and pulled out other interacting molecules. In vivo and in vitro protein-protein interaction of these molecules with ARL1 and other ARLs will be used to examine their specific interactions. [2] To characterize the structure and function of ARL1 protein.
We have demonstrated ARL1, similar to ARFs, is N-myristoylated in vitro. We will determine whether this modification is involved in membrane anchoring or other biological activity. We will define the membrane anchoring activity in vivo. The role of ARL1 may have clues by overexpressing its dominant negative mutants (ARL1T31N and ARL1Q71L). We will establish
5
stable transfectants expressing these mutants and determine their effects on translocation of lysosome-specific-marker proteins in these transfectants. GTPγS binding and CTA-catalyzed ADP-ribosyltransferase activities of ARL1 and its mutants will also be examined. Expression and localization of endogenous ARL1 protein in cells will be examined. Electronic microscopy will also be performed to more define its subcellular localization.
[3] To study the molecular mechanisms of ARL1 interacted with its effectors.
Dominant negative mutants of ARL1 and its interacting proteins will be tested whether they can affect Golgi -lysosome trafficking. We will also examine whether ARL1 and its interacting proteins can shuttle between trans-Golgi and lysosome. Identified hARL1-complex proteins will be used to identify their specific transport cargos. We will further examine the hARL1-complex in Golgi-lysosome sorting function. Sorting signal and binding partners of the ARL1 regulated transport vesicles will also be examined.
6 3. Progress Summary
Summarize concisely the results obtained for each specific aim during the past year (or reporting period). Negative results, if any, should also be included and approaches taken to improve the prospects of the project discussed. (Do not exceed 5 pages, not including figures and references.)
Results
Identification of molecules that might act as down-stream effectors of hARL1
For further understanding the possible function of hARL1, we used the yeast two-hybrid screen to identify molecules that might act as binding partners of hARL1. The putatively constitutively active mutant of human ARL (hARL1Q71L) fused at their amino-terminus with the DNA binding domain of LexA as bait to screen human fetal liver cDNA library (Clontech). The bait caused no intrinsic transcriptional activation of the reporters. We get about 43 positive clones from 1.2 x 106 transformants. The DNA sequence of each library insert was determined and summary to nine proteins. They include 9 clones of Golgin-245(accession number U31906), 9 clones of Apolipoprotein C-Ⅱ(ApoCⅡ)(accession number NM_000483), 4 clones of Arfaptin 2 (Partner of RAC1) (POR1 protein) (accession number P53365), 2 clones of Apolipoprotein B(ApoB)(accession number AH003569), one clone of brain secretory protein hSec10p (HSEC10) (accession number U85946), and one clone of calcium and integrin binding 1 (calmyrin) (CIB1)( accession number NM_006384). Furthermore the human ARL1 complex was identified by tandem affinity purification method coupling with two-dimensional electrophoresis and mass spectrometry identification of unknown protein complex components. The key feature of this technology is the use of two different affinity purification tags that are fused to variant human ARLs and mutated human ARL1 by genetic methods. Performing two consecutive purification steps using affinity purification tags that have gentle washing and elution conditions allows for isolation without disrupting the targeted complex. Recently we defined at least four interacting protein spots for human ARL1Q71L protein complex (figure 17). There were two protein spots with molecular masses approx. of 20 kDa and pI approx. of 4.5 identified as myosin regulatory light chain 2 (accession number P19105) and myosin regulatory light chain MRCL3 (accession number NP_006462), respectively (figure 17, spots 1 and 2). We will be further characterized their interactions if having functional significant.
hARL1 specifically interacts with Golgin 245 at its C-terminal GRIP domain
The clones of Golgin-245 which we get from library screen were encoded amino acids range from 1846 to 2229. The longest fragment of Golgin-245 clones (amino acid 1846-2229) was named Golgin-245C’. Thus, the interaction of Golgin-245 and ARL1 appeared to involve its C terminus. We also used yeast two-hybrid assay to identify whether the interaction between Golgin-245 and hARL1 is nucleotide-dependent and specificity. Yeast transformants containing
7
interacting proteins that transactivate two reporter genes, HIS3 and LacZ, exhibit β-galactosidase activity and can grow on minimal medium lacking histidine. As illustrated in Table 2 and Figure 1, LexA-hARL1Q71L, but not LexA-hARL1T31N, LexA-hARL1, LexA-hARL2, LexA-hARL3, LexA-hARL4, LexA-hARL4Q79L, LexA-hARL5, or LexA-hARL5Q80L, can interacted with the Gal4AD-Golgin-245C fusion protein and activated the reporter genes. So Golgin-245 does not interact with class II of ARL (ARL2 and ARL3) and class Ⅲ of ARL (ARL4 and ARL5). The interaction is specific to ARL1 and nucleotide-dependent.
Interestingly, when we delete the N-terminal 17 amino acid of hARL1, either hARL1 Q71Ld17N or hARL1WTd17N can interact with Golgin-245C’. It should be noted that positively charged residues in the N terminus have been postulated to be important for membrane binding by participating in electrostatic interactions with the lipid bilayer (Amor et al., 1994; Antonny et al., 1997). We suggest hARL1(WT, d17N) can interact with Golgin-245C may be the result of conformation effect of the deleted N-terminal residues 1-17 of hARL1.
The GRIP domain of Golgin-245 is included in amino acid 1846-2229. To map the interaction region for hARL1 in Golgin-245 if involve the GRIP domain, we construct the deletion mutants of Golgin-245C’. They are Golgin-245C’dC (amino acid 1846-2032) and Golgin-245C’dN (amino acid 2033-2229) (Fig.1A). The results from our yeast two-hybrid assay showed that the GRIP domain in Golgin-245 is essential for hARL1 association (Fig.1B).
We also performed GST pull-down experiments to confirm the results with the yeast two-hybrid assay. Recombinant His-hARL1 and its His fusion mutants were incubated with GST-Golgin-245C’, GST-Golgin-245C’dC or GST-Golgin-245C’dN immobilized on glutathione Sepharose. As shown in Figure 2, only fragment of the recombinant GST-Golgin-245 containing GRIP domain, but not GST, associate with the hARL1 and hARL1Q71L. His-ARL1T31N and His-ARL4 don’t interact with GST-Golgin-245. Base on the in vitro pull down assay hARL1WT can direct interact with Golgin-245. It was different from the result by yeast two-hybrid assay. We assume that it maybe lack post-translation modification of ARL1 in bacteria result in ARL1 prefer GTP-bound form in bacteria. Meanwhile, we observe that one smaller protein fragment exist in our reaction solution with stronger interaction to Golgin-245. This protein fragment could be detected by our ARL1 antibody, but not by His antibody, suggest it may be N-terminal degraded form of hARL1. Extract the protein fragment from acrylamide gel and sequence by Edman degradation show it was the deleted N –terminal residues 1-17 of ARL1.
Cross-reaction between human ARL1 and yeast ARL1
The homologue of Human ARL1 in yeast S. cerevisiae is yARL1. The genome of the yeast S.
cerevisiae encodes only one protein with a GRIP domain, namely the coiled-coil protein Imh1p.
The IMH1 gene was originally isolated as a suppressor of the temperature-sensitive lethality of a mutation in the small GTP-binding protein Ypt6p, a close relative of the mammalian protein Rab6 (Li et al., 1996). Using yARL1QL as bait to screen yeast cDNA library we obtained Imh1p (Liou
8
GRIP domain, we want to known whether Golgin-245 interact to yARL1 and vice versa. Identified by yeast two-hybrid assay, only GTP bound form of ARL1, either human or yeast, can interact with GRIP domain-containing Golgin-245 or Imh1p (Fig.). So we suggest that we can use yARL1 as a model in yeast system to further investigate the physiological function of ARL1.
ARF effector arfaptin-2/POR1 can interact with ARL1
Using hARL1Q71L as bait to screen library, one of the interacting proteins obtained is arfaptin 2/POR1, as previous finding (Hillary et al., 1999; Lu et al., 2002). We used yeast two-hybrid assay to further confirm the specificity of interaction between ARL1 and arfaptin 2. VP16-arfaptin2 interacted with LexA-hARL1WTd17N, LexA-hARL1Q71L and LexA-hARL1Q71Ld17N, but not LexA-hARL1T31N, LexA-hARL1, LexA-hARL2, LexA-hARL3, LexA-hARL4, LexA-hARL4Q79L, LexA-hARL5, or LexA-hARL5Q80L. Like golgin-245, the interaction of arfaptin 2 and ARL1 was specific and depended on nucleotide. Arfaptin 2 also interacted to ARF1Q71L as previous finding (Kanoh et al., 1997). We also test the interaction of arfaptin 1 and ARL1. Surprisingly, Arfaptin 1 can interact with hARL1Q71L weakly, but not interact with hARL1Q71Ld17N.
Other possible binding partners of hARL1
Two of hARL1 possible interacting partner are ApoC2 and ApoB by our yeast two-hybrid library screen. Apolipoproteins and their receptors are the main controllers of lipid metabolism. Human ApoCs are proteinconstituents of chylomicrons, VLDL, and HDL. Apolipoprotein C-II (ApoC2) is a necessary cofactor for the activation of lipoprotein lipase, the enzyme that hydrolyzes triglycerides in plasma and transfers the fatty acids to tissues. The importance of ApoC2 has unequivocally been demonstrated in patients with geneticdefects in the structure or production of apoC2, all of whomdisplay high circulating levels of triglycerides (TGs) and are phenotypically indistinguishable from patients with LPL deficiency (John et al., 1999). ApoC2 is synthesized with a 22-residue signal peptide that is cleaved cotranslationally in the rough endoplasmic reticulum (Sharpe et al., 1984). The remaining single polypeptide chain of 79 amino acid residueshas a calculated molecular mass of 8.8 kDa (Fojo et al., 1984). The structureof ApoC2 is predicted to contain 3 helical regions which are thought to be involvedin phospholipid binding (Captano et al., 1979; Kinnunen et al., 1977). To test whether ApoC2 specifically interact with hARL1, not to other ARF and ARL, we performed yeast two-hybrid assay. Unlike Golgin-245 and arfaptin 2, Gal4 AD-ApoC2 only interacted with LexA-hARL1(Q71L), but not with LexA-hARL1, LexA-hARL1(d17N), LexA-ARL1(d17N, Q71L), LexA-hARL1(T31N), LexA-hARL2, LexA-hARL3, LexA-hARL4, LexA-hARL4(Q79L), LexA-hARL5, LexA-hARL5(Q80L), LexA-hARF1 or LexA-hARF6. It suggests that the residues 1-17 of ARL1 may be required for hARL1 and ApoC2 association and their interaction is nucleotide-dependent. We also assume that the N terminal region of hARL1 isn’t essential for Golgin-245 and arfaptin 2/POR1. When deleted N terminalresidues 1- 17 of ARL1, either hARL1WTd17N or
9
hARL1Q71Ld17N have stronger interaction than full-length hARL1WT and hARL1Q71L. We will further investigate the significance of interaction between hARL1 and its partners.
Characterization of antibodies recognizing hARL1, golgin245 or arfaptin2
In order to examine whether golgin245 and arfaptin2 are functionally related to Arl1, we used peptide (hARL1B) or recombinant proteins (GST-golgin245C’ and GST-arfaptin2) as antigens to immunize rabbits. Anti-hARL1, anti-golgin245 and anti-arfaptin2 antibodies could recognize recombinant His-hARL1, GST-golgin245C’ and GST-arfaptin2, respectively. By indirect immunostaining probed with these antibodys, perinuclear Golgi-like staining could be detected in COS-b cells. Antiserum preincubated with its antigen but not control lost its staining signal.
Arl1(T31N) is a dominant negative mutant
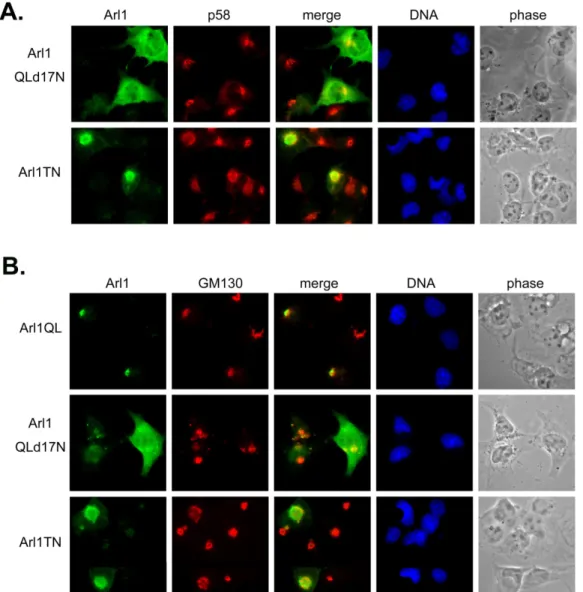
Constitutively inactive mutant of Arl1, Arl1 (T31N) generated by substituting Thr at position 31 with Asn are regarded as a dominant negative form and disassemble Golgi apparatus (Lu et al., 2001). Overexpressed hARL1(T31N) formed a perinuclear structure and indeed altered the pattern of examined Golgi proteins containing GM130. This might suggest that hARL1(T31N), like hARF1(TN), overall affects the Golgi apparatus and dues to disperse golgi proteins without specificity.
Expression of Arl1(d17N,Q71L) disperses golgin245
Under morphological assay, both GTP-restricted Arl1 and ARF1 expand Golgi apparatus (Lu
et al., 2001; Van Valkenburgh et al., 2001). It seems that Arl1, like ARF1, also involves in the
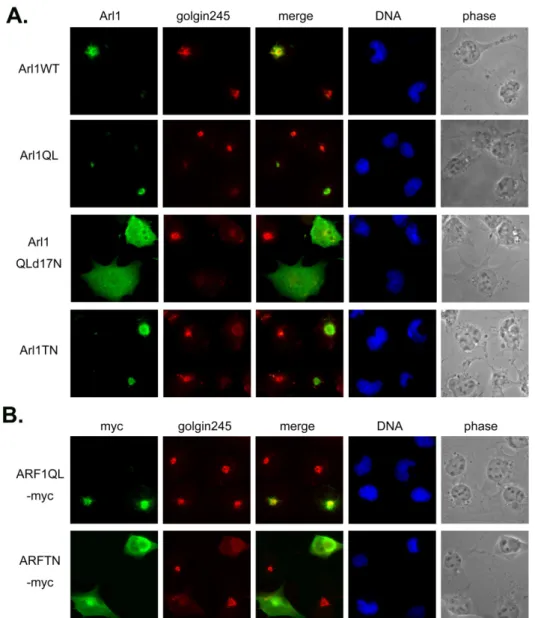
functions of Golgi. Arl1 enriches on the trans side of the Golgi (Lu et al., 2001) and hArl1 is myristoylated at the second residue, Gly (Lee et al., 1997). A preferencially GTP-bound form of hArl1 termed hArl1(Q71L) was generated by replacing Qln at nucleotide 71 with Leu. Extopically expressed hArl1(Q71L) localized to Golgi marked by GM130 and specifically decreased the staining signal of golgin245. It’s ambiguous that the active form of Arl1 is predicted to recruit its effectors and more resistants to BFA treatment (Lu et al., 2001; Lu and Hong, 2003). However, according to our result, Arl1(Q71L) not only didn’t recruit golgin245 but also interfered the Golgi targeting of golgin245; hARF1(QL) as a control didn’t affect golgin245. One interpretation could be that a part of epitopes of golgin245 were masked because of interacting with Arl1(Q71L). We could not detect the interaction between hArl1(Q71L) and the GRIP domain of golgin245 (see below), though Arl1(Q71L)-EGFP was reported to precipitate golgin245 (Lu and Hong, 2003). How hArl1(Q71L) and golgin245 mutual influence should be study further.
Arl1(d17N, Q71L) created by deleting the N-terminal residues 1-17 of Arl1(Q71L) dispersed evenly in nucleoplasm and cytoplasm when expressed in COS-b cells. Without the myristoylation site, hArl1, even the active form, preferred to resident at cytosol. We can’t distinguish the
10
hArl1(d17N,Q71L) and endogenous hArl1, so whether endogenous hArl1 also localizes at Golgi is not clear. It’s interesting that golgin245 was totally scattered from Golgi in cells expressed hArl1(d17N,Q71L). Overexpressed hArl1(d17N,Q71L) didn’t affect GM130, β-COP and p58 and affected golgin97 very less, if there was any effect. To address whether those effects of hArl1 was through direct interaction with golgin245; immunoprecipitation could give us a hint. Because the expression level of untagged hArl1 mutants were too low to be detected by western blot, we tagged the TAP at the C terminal of hArl1 to generate hArl1-TAP mutants whose subcellualr localization were the same as untagged hArl1 mutants. On the other hand, a construct containing the C-termial residues 2033-2229 having GRIP domain of golgin245 was inframe fused with a Flag tag at N terminus, termed Flag-golgin245C’dN. 293T cells were cotransfected with hArl1-TAP and Flag-golgin245C’dN and subjected to pull down with protein A-beads. Flag-golgin245C’dN was coprecipitated with hArl1(d17N,Q71L)-TAP but not with hArl1(Q71L)-TAP or hArl1(T31N)-TAP. The N-terminal extensions of Arf family proteins are predicted to form amphipathic helices; in GDP-bound Arf1 and Arf6, the interswitch is retracted and forms a pocket to which the N-terminal helix binds, the latter serving as a molecular hasp to maintain the inactive conformation (Pasqualato et al., 2002; and references in this article). This consists with the precipitation result; without the N-terminal extantion, hArl1(Q71L)-TAP could mimic the active form of hArl1better than hArl1(Q71L) could. An easy interpretation is that because hArl1(d17N,Q71L) interacts with golgin245 and residents at cytosol, it interferes with Golgi targeting of golgin245.
Expression of Arl1(d17N,Q71L) disperses arfaptin2
Base on the same approach, the relationship between hArl1 and arfaptin2 was examined. Because both antibodies for detecting hArl1 and arfaptin2 were raised by rabbit, the two signals can’t be detected by immunofluorescence at the same time. Flag-arfaptin2 was constructed by fussion the Flag tagged at the N terminus with arfaptin2. It localized to Golgi as reporting (Frantz et al., 2002; Peters et al., 2002) when expression level was medial; it diffused to cytosol when expression level was high. COS-b cells were cotransfected with Flag-arfaptin2 and hArl1 mutants then subjected to immunostaining. Flag-arfaptin2 couldn’t target to Golgi in cells expressing hArl1(d17N,Q71L). HArl1(Q71L) didn’t change the localization of Flag-arfaptin2. We also single transfected COS-b cells with the C-terminal myc-tagged hARL1(d17N, Q71L); the signal of endogenous arfaptin 2 was disappeared in cells expressing hARL1(d17N, Q71L)-myc. These results suggest that arfaptin 2 is an effector of hARL1. Again, 293T cells were cotransfected with Flag-arfaptin2 and hArl1-TAP mutants and subjected to pull down with protein A-beads. Non of hArl1-TAP mutants could precipitate Flag-arfaptin2.
GDP-bound form of Arl1 colocalizes with lysozome
11
vesicles transported between Golgi and plasma membrane. After cells were transfected with Arl1(T31N)-myc then processed to double labeling with anti-myc antibody and lysotracker. The signal of Arl1(T31N) and lysotracker partially overlapped.
Discussion
The GTP-restricted form of Arl1, Arl1 (Q71L), is thought to recruit its effectors (Lu and Hong, 2003) and arrests the transport of VSV-G from Golgi to cell surface; VSV-G transport wasn’t abolished in cells overexpressing Arl1(G2A, Q71L) lacking the myristoylation site (Lu et
al., 2001). These data suggest that myristoylated Arl1 (Q71L) prefers to resident at Golgi and
retains its effectors; then blocks the vesicular transport from the trans Golgi network (TGN). On the other hand, the Golgi-targeting of golgin 245 and golgin 97 is reported to depend on Arl1 (Lu and Hong, 2003). But this proposal still is a puzzle; in our observation, hArl1 (Q71L) seems to interfere Golgi targeting of golgin245. There are other results disagreement with the literature. Without the myristoylation site of Arl1 (Q71L), Arl1 (G2A, Q71L) loses the dominant effect caused by Arl1(Q71L) (Lu et al., 2001). But hArl1 (d17N,Q71L) enhances the effect caused by hArl1(Q71L). A myc tag was inframe fused to the C terminus with hArl1 and the effect of this fusion protein, hArl1(d17N, Q71L)-myc, was the same as untagged mutant. To specify the effect of hArl1(d17N, Q71L)-myc, hArl1(G2A)-myc was produced by replacing Gly at position 2 with Ala of hArl1-myc and it was detected in the cytosol and nucleoplasm without Golgi enrichment. Although hArl1(G2A) and hArl1(d17N, Q71L) had the same distribution, hArl1(G2A) totally didn’t scatter golgin245 or arfaptin2 (data not shown).
Although arfaptin 2 is a common effector of Arl1 and ARF1, the significance of these relationship is unclear. Arfaptin2 was reported to interact with Arl1 to increase GTPγS binding in vitro (Van Valkenburgh et al., 2001). We wondered if arfaptin2 could prevent hArl1 from diffusion caused by BFA treatment. The answer was no; hArl1 diffused by BFA treatment even in cells overexpressing arfaptin 2 (data not shown). Arfaptin 2 is not likely to increase GTP binding of hArl1 in vivo. There are some data about arfaptin 1. By using in vitro translocation study, arfaptin1 in HL-60 cell cytosol is recruited by ARF to Golgi membrane in a GTPγS-dependent and brefeldin A-sensitive manner (Exton et al., 1997). In addition, arfaptin1 is a potent inhibitor of ARF actions on cholera toxin and phospohlipase D (Kanoh et al., 1998).
Some studies in yeast imply Arl1 involving in the transport from TGN to vacuoles. Morphological and biochemical analyses of cdc28 dlp2 (a single mutation in the gene ARL1) suggested that a defect in central vacuole formation is caused by aberrant membrane trafficking, although the protein-sorting to vacuoles is not affected (Abudugupur et al., 2002). arl1△ trains
secreted less protein, mislocalized carboxypeptidase Y to the plasma membrane and uptaked less fluidphase marker (Rosenwald et al., 2002). Whether hArl1 regulates the transport to lysosome or exocytosis should be studied further. Base on the advantage of that hArl1(d17N, Q71L)
12
specifically mislocalized golgin245 and arfaptin2; the proteins in the cascade regulated by hArl1 should be affected. We will examine the staining pattern of endosomal protein EEA1 in cells expressing hArl1(d17N, Q71L).
How is sorting signal recognition regulated so that interaction with each vesicle-receptor binding protein occurs only at the appropriate organelle? Cooperative association of cargo, vesicle binding proteins, and additional factors into mutimeric, trans-Golgi and/or lysosome-specific complexes will be investigated in this project.
References
Amor, C., Harrison, D., Kahn, R., and Ringe, D. (1994) Structure of the human ADP-ribosylation factor 1 complexed with GDP. Nature 372, 704-708.
Antonny, B., Beraud-Dufour, S., Chardin, P., and Chabre, M. (1997) N-Terminal hydrophobic residues of the G-protein ADP-ribosylation factor-1 insert into membrane phospholipids upon GDP to GTP exchange. Biochemistry 36, 4675-4684.
Abudugupur, A., Mitsui, K., Yokota, S., and Tsurugi, K. (2002) An ARL1mutation affected autophagic cell death in yeast, causing a defect in central vacuole formation. Cell. Death Differ. 9, 158-168.
Bhamidipati, A., Lewis, S. A. and Cowan, N. J. (2000) ADP ribosylation factor-like protein 2 (Arl2) regulates the interaction of tubulin-folding cofactor D with native tubulin. J. Cell Biol. 149, 1087–1096.
Boman, A. L., and Kahn, R. A. (1995) Arf proteins: the membrane traffic police? Trends Biochem.
Sci. 20, 147-150.
Brown, D. L., Heimann, K., Lock, J., Kjer-Nielsen, L., van Vliet, C., Stow, J. L., Gleeson, P. A. (2001) The GRIP domain is a specific targeting sequence for a population of trans-Golgi network derived tubulo-vesicular carriers. Traffic. 2, 336-44.
Captano, A. L., Kinnunen, P. K. J., Breckenridge, W. C., Gotto, A. M. Jr., Jackson, R. L., Little, J. A., Smith, L. C., Sparrow, J. T. (1979) Lipolysis of apoC-II deficient very low density lipoproteins: enhancement of lipoprotein lipase action by synthetic fragments of apoC-II. Biochem. Biophys.
13
Res. Commun. 89, 951–957.
Cowan, D.A., Gay, D., Bieler, B. M., Zhao, H., Yoshino, A., Davis, J.G., Tomayko, M. M., Murali, R., Greene, M. I., Marks, M. S. (2001) Characterization of mouse tGolgin-1 (golgin-245/trans-golgi p230/256 kD golgin) and its upregulation during oligodendrocyte development. DNA Cell Biol. 21, 505-17.
D’Souza-Schorey, C., Boshans, R. L., McDonough, M., Stahl, P. D. and Van Aelst, L. (1997) A role for POR1, a Rac1-interacting protein, in ARF6-mediated cytoskeletal rearrangements. EMBO
J. 16, 5445-5454.
Dell’Angelica, E. C., Puertollano, R., Mullins, C., Aguilar, R. C., Vargas, Lowe, S. L., Wong, S. H. and Hong, W. (1996) The mammalian ARF-like protein 1 (Arl1) is associated with the Golgi complex. J. Cell Sci. 109, 209-220.
Erlich, R., Gleeson, P. A., Campbell, P., Dietzsch, E., Toh, B.-H. (1996) Molecular characterization of trans-Golgi p230: a human peripheral membrane protein encoded by a gene on chromosome 6p12-22 contains extensive coiled-coil alpha-helical domains and a granin motif. J.
Biol. Chem. 271, 8328-8337.
Frantz, C., Coppola, T., and Regazzi R. (2002) Involvement of Rho GTPases and Their Effectors in the Secretory Process of PC12 Cells. Exp. Cell Res. 273, 119–126.
Fojo, S. S., Law, S. W., Brewer, H. B. Jr. (1984) Human apolipoprotein C-II: complete nucleic acid sequence of preapolipoprotein C-II. Proc Natl Acad Sci U S A. 81, 6354–6357.
Fritzler, M. J., Lung, C.-C., Hamel, J. C., Griffith, K. J., Chan, E. K. L. (1995) Molecular characterization of golgin-245, a novel Golgi complex protein containing a granin signature. J.
Biol. Chem. 270, 31262-31268.
Hirst, J., and Robinson, M. S. (1998) Clathrin and adaptors. Biochim. Biophys. Acta 1404, 173-193.
Hong, J.-X., Lee, F-J. S., Patton, W. A., Lin, C.-Y., Moss, J., and Vaughan, M. (1998) Phospholipid- and GTP-dependent activation of cholera toxin and phospholipase D by human ADP-ribosylation factor-like protein 1 (HARL1). J. Biol. Chem. 273, 15872-15876.
Joneson, T., McDonough, M., Bar-Sagi, D., and Van Aelst, L. (1996) Rac regulation of actin polymerization and proliferation by a pathway distinct from Jun kinase. Science 274, 1374–1376.
14
Jong, M. C., Hofker, M. H., Havekes, L. M. (1999) Role of ApoCs in lipoprotein metabolism: functional differences between ApoC1, ApoC2, and ApoC3. Arterioscler Thromb Vasc Biol. 19, 472-484.
Kanoh, H., Williger, B.-T., Exton, J. H. (1997) Arfaptin 1, a putative cytosolic target protein of ADP-ribosylation factor, is recruited to Golgi membranes. J. Biol. Chem. 272, 5421-5429.
Kinnunen, P. K. J., Jackson, R. L., Smith, L. C., Gotto, A. M. Jr., Sparrow, J. T. (1997) Activation of lipoprotein lipase by native and synthetic fragments of human plasma apolipoprotein C-II. Proc
Natl Acad Sci U S A 74, 4848–4851.
Kjer-Nielsen, L., Teasdale, R. D., van Vliet, C., Gleeson, P. A. (1999) A novel Golgi-localisation domain shared by a class of coiled-coil peripheral membrane proteins. Curr Biol. 9, 385-388. Kooy, J., Toh, B.-H., Pettitt, J. M., Erlich, R., Gleeson, P. A. (1992) Human autoantibodies as reagents to conserved Golgi components: characterization of a peripheral, 230-kDa compartment-specific Golgi protein. J. Biol. Chem. 267, 20255-20263.
Lee, F. J., Huang, C. F., Yu, W. L., Buu, L. M., Lin, C. Y., Huang, M. C., Moss, J. and Vaughan, M. (1997) Characterization of an ADP-ribosylation factor-like 1 protein in Saccharomyces cerevisiae.
J. Biol. Chem. 272, 30998-31005.
Li, B. J., and Warner, J. R. (1996) Mutation of the Rab6 homologue of Saccharomyces cerevisiae, YPT6, inhibits both early Golgi function and ribosome biosynthesis. J. Biol. Chem. 271, 16813-16819.
Lin C.-Y., Huang P.-H., Liao W.-L., Cheng H.-J., Huang C.-F., Kuo J.C., Patton W.A., Massenburg D., Moss J., and Lee F.-J. S. (2000) ARL4, an ARF-like Protein that is Developmentally Regulated and Localized to Nuclei and Nucleoli. J. Biol. Chem. 275, 37815-37823.
Lin C.-Y.,. Li C.-C., Huang P.-H., and Lee F.-J. S. (2002) A Developmentally Regulated ARF-like protein (ARL5), localized to Nuclei and Nucleoli, Is Interacted with Heterochromatin Protein 1. J.
Cell Sci. 115, 4433-4445.
Lippincott-Schwartz, J., Cole, N. B., and Donaldson, J. G. (1998) Building a secretory apparatus: role of ARF1/COPI in Golgi biogenesis and maintenance. Histochem. Cell Biol. 109, 449-462. Lu, L., Horstmann, H., Ng, C., and Hong, W. (2001) Regulation of Golgi structure and function by
15 ARF-like protein 1 (Arl1). J Cell Sci. 114, 4543-4555.
McConville, M. J., Ilgoutz, S. C., Teasdale, R. D., Foth, B. J., Matthews, A., Mullin, K. A., Gleeson, P. A. (2002) Targeting of the GRIP domain to the trans-Golgi network is conserved from protists to animals. Eur. J. Cell Biol. 81, 485-495.
Moss, J., and Vaughan, M. (1998) Molecules in the ARF orbit. J. Biol. Chem. 273, 21431-21434. Munro, S., and Nichols, B. J. (1999) The GRIP domain - a novel Golgi-targeting domain found in several coiled-coil proteins. Curr Biol. 9, 377-380.
Pasqualato, S., Renault, L., and Cherfils, J. (2002) Arf, Arl, Arp and Sar proteins: a family of GTP-binding proteins with a structural device for ‘front–back’ communication. EMBO Rep. 3, 1035-1041.
Peters, P. J., Ning, K., Palacios, F., Boshans, R. L., Kazantsev, A., Thompson, L. M., Woodman, B., Bates, G. P., D'Souza-Schorey, C. (2002) Arfaptin 2 regulates the aggregation of mutant huntingtin protein. Nature Cell Biol. 4, 240-245.
Rosenwald, A.G., Rhodes, M. A., Van Valkenburgh, H., Palanivel, V., Chapman, G., Boman, A., Zhang, C.-J., and Kahn, R. A. (2002) ARL1 and membrane traffic in Saccharomyces cerevisiae.
Yeast 19, 1039–1056.
Sharpe, C. R., Sidoli, A., Shelley, C. S., Lucero, M. A., Shoulders, C. C., Baralle, F. E. (1984) Human apolipoproteins AI, AII, CII and CIII, cDNA sequences and mRNA abundance. Nucleic
Acids Res. 12, 3917–3932.
Shin, O.-H., and Exton, J. H. (2001) Differential Binding of Arfaptin 2/POR1 to ADP-Ribosylation Factors and Rac1. BBRC 285, 1267–1273.
Tamkun, J. W., Kahn, R. A., Kissinger, M., Brizuela, B., Rulka, C., Scott, M. P. and Kennison, J. A. (1991) The arflike gene encodes an essential GTP-binding protein in Drosophila. Proc. Natl.
Acad. Sci. 88, 3120-3124.
Tarricone, C., Xiao, B., Justin, N., Walker, P. A., Rittinger, K., Gamblin, S. J., Smerdon, S. J. (2001) The structural basis of Arfaptin-mediated cross-talk between Rac and Arf signalling pathways.
Nature 411, 215-219.
16
Effects of Arfaptin 1 on Guanine Nucleotide-dependent Activation of Phospholipase D and Cholera Toxin by ADP-ribosylation Factor. J. Biol. Chem. 273, 20697-20701.
Van Aelst, L., Joneson, T., Bar-Sagi, D. (1996) Identification of a novel Rac1-interacting protein involved in membrane ruffling. EMBO J. 15, 3778-3786.
Van Valkenburgh, H., Shern, J. F., Sharer, J. D., Zhu, X., Kahn, R. A. (2001) ADP-ribosylation factors (ARFs) and ARF-like 1 (ARL1) have both specific and shared effectors: characterizing ARL1-binding proteins. J. Biol. Chem. 276, 22826-37.
Williger, B. T., Ostermann, J., and Exton, J. H. (1999) Arfaptin 1, an ARF-binding protein, inhibits phospholipase D and endoplasmic reticulum/Golgi protein transport. FEBS Lett. 443, 197-200.
17
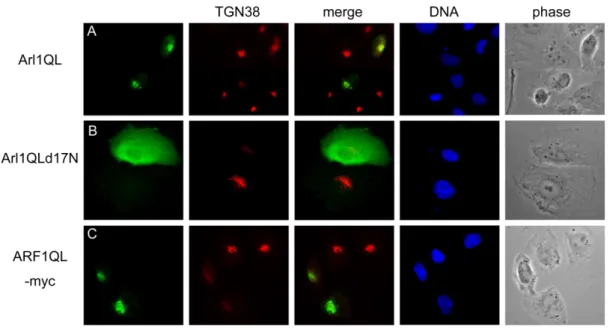
Fig 1. Constitutively active Arl1, termed Arl1QL interferes the Golgi association of golgin245. Cos-b cells were transiently transfected with constructs expressing (A) Arl1wt or Arl1QL or Arl1QLd17N or Arl1TN or (B) ARF1QL-myc or ARF1TN-myc. Cells were fixed and double labeled with a monoclonal antibody against golgin245 and a limiting amount of Arl1 antibodies to reveal the exogenous expressed Arl1 (A), or a antibody against myc tag to detect myc-tagged ARF1. Only Arl1QL but not ARF1QL-myc affects the localization of golgin245. Arl1QL without the N terminal 17 residues evenly diffuses in cells and totally scatters golgin245. The inactive forms, Arl1TN and ARF1TN-myc seem to overall change the Golgi structure.
18
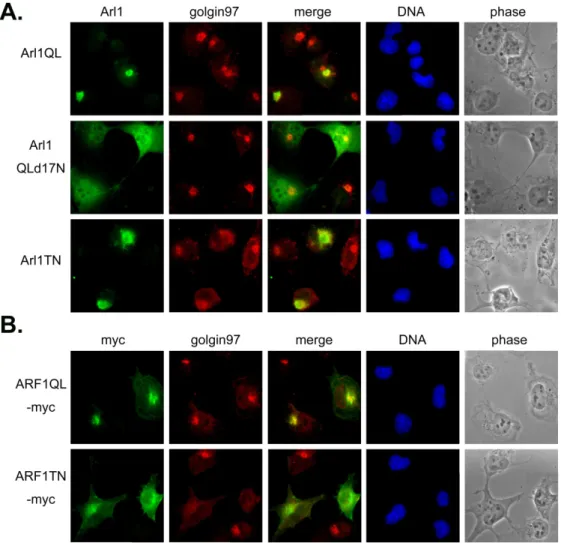
Fig2 . Arl1QL has no effect on the GRIP domain containing protein, golgin97. Cos-b cells were transiently transfected with constructs expressing (A) Arl1QL or Arl1QLd17N or Arl1TN or (B) ARF1QL-myc or ARF1TN-myc. Cells were fixed and double labeled with a monoclonal antibody against golgin97 and a limiting amount of Arl1 antibodies to reveal the exogenous expressed Arl1 (A), or a antibody against myc tag to detect myc-tagged ARF1. Neither Arl1QL nor Arl1QLd17N changes the staining pattern of golgin97. The inactive forms, Arl1TN and ARF1TN-myc diffuse golgin97.
19
Fig 3. Arl1QL and Arl1QLd17N do not affect the cis Golgi protein β-COP. Cos-b cells were transiently transfected with constructs expressing (A) Arl1QL or Arl1QLd17N or Arl1TN or (B) ARF1QL-myc or ARF1TN-myc. Cells were fixed and double labeled with a monoclonal antibody against β-COP and a limiting amount of Arl1 antibodies to reveal the exogenous expressed Arl1 (A), or a antibody against myc tag to detect myc-tagged ARF1.
20
Fig 4. The inactive form of Arl1 disassembles the Golgi apparatus. Cos-b cells were transiently transfected with constructs expressing Arl1QL or Arl1QLd17N or Arl1TN. Cells were fixed and double labeled with a monoclonal antibody against p58 (A) or GM130 (B) and a limiting amount of Arl1 antibodies to reveal the exogenous expressed Arl1. Only the inactive form but not the active form of Arl1 changes the Golgi marked by p58 (A) or GM130 (B).
21
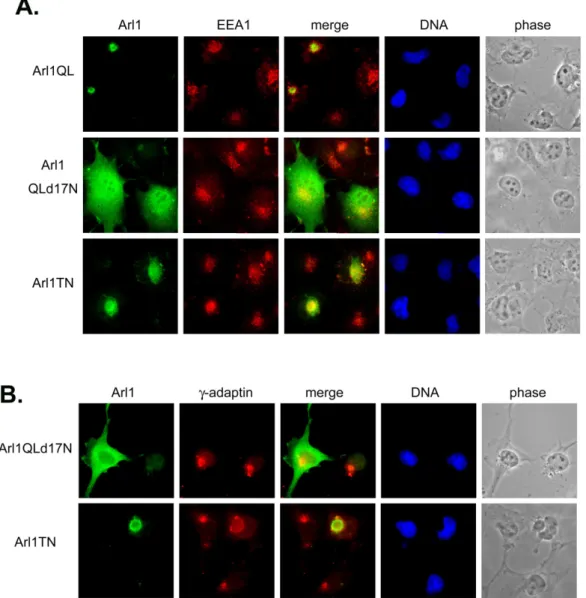
Fig 5. The transport pathway from golgi to endosome seems not to be regulated by Arl1. Cos-b cells were transiently transfected with constructs expressing Arl1QL or Arl1QLd17N or Arl1TN. Cells were fixed and double labeled with a monoclonal antibody against EEA1 (A) or γ-adaptin (B) and a limiting amount of Arl1 antibodies to reveal the exogenous expressed Arl1. Arl1QL and Arl1QLd17N do not affect the endosomal protein EEA1 and the adaptor protein, γ-adaptin involved in Golgi-endosome transport. The inactive form of Arl1, Arl1TN has a nonspecific effect.
22
Fig 6. The TGN38 shuttling between Golgi and plasma membrane seems to be affected by Arl1 and ARF1. Cos-b cells were transiently transfected with constructs expressing Arl1QL (A) or Arl1QLd17N (B) or ARF1QL-myc (C). Cells were fixed and double labeled with a monoclonal antibody against TGN38 and a limiting amount of Arl1 antibodies to reveal the exogenous expressed Arl1 (A-B) or an anti-myc antibody to detect C-terminal myc-tagged ARF1QL (C).
23
Fig 7. Characterizing anti-Arl1 antibody. Anti-Arl1 antibody was pre-incubated with (A) PBS only or (B) 1µg recombinant Arl1 or (C) Arl1 peptide which was used as antigen to generate this antibody. Cos-b cells were fixed and probed with these pre-incubated antibodies. The staining signal can be blocked by the recombinant protein (B) and the peptide (C).
24
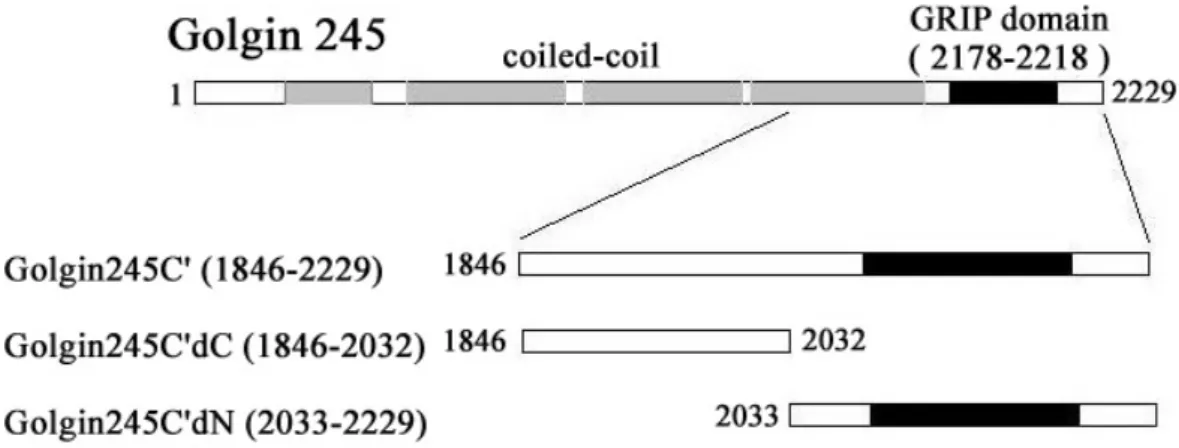
Fig 8. The schematic of golgin245. Golgin245 contains the coiled-coil region (gray) and the GRIP domain (black). Golgin245C’ is the interaction clone from yeast two hybrid screen using Arl1QL as bait.
Fig 9. Arl1QLd17N precipitates the C terminal region of golgin245. 293T cells were transient transfected with the C terminal TAP fused Arl1 mutants and Flag-tagged golgin245C’dN (lane 1-3, 7-9) or Flag-tagged full length arfaptin2 (lane 4-6, 10-12). Cells were then subjected to precipitate with IgG beads and the Arl1-TAP complexes were pelleted. The precipitate complexes were solved by SDS-PAGE and probed with anti-TAP antibody to detect Arl1-TAP (lane 1-6) or with anti-Flag antibody to detect Flag-golgin245C’dN (lane 7-9) or Flag-arfaptin2 (lane 10-12).
25
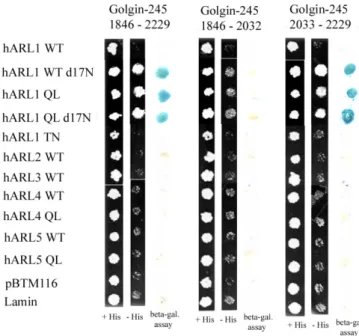
Fig 10. Arl1QLd17N interacts with golgin245C’ in vitro. Recombinant His tagged Arl1QL or Arl1QLd17N or GST-fused golgin245C’ were purified respectively. 10 µg recombinant Arl1QL was incubated with GST-golgin245C’ (lane 5-7) or GST (3) and then was subjected to GST-pull down assay. Not only the predicted size of Arl1QL but also the smaller size one associate with golgin245C’. These association can be competed by ARL1QLd17N (lane 5-8).
Fig 11. The C terminal region containing GRIP domain of golgin245 interacts with Arl1 but not inactive Arl1. Different constructs of golin245 were transformed into yeast AMR70 and mate with yeast strain L40 transformed with one of the small GTPase as indicated. Beta-gal assays were performed to dissect the interactions.
26
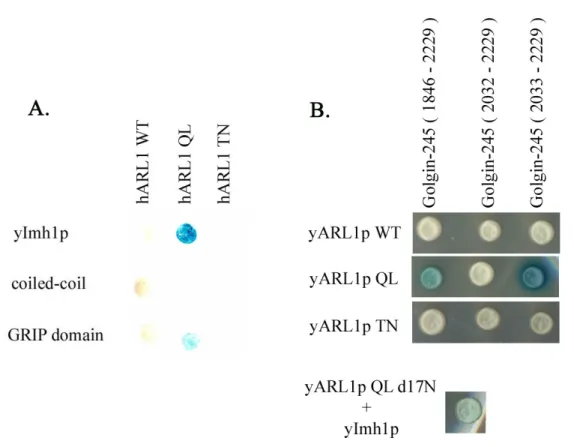
Fig 12. human and yeast Arl1 can cross talk in yeast two hybrid assay. Human Arl1 interacts with yeast Imh1p through its GRIP domain (A). Yeast Arl1p interacts with human golgin245.
27
Fig 13. arfaptin2 is a common effector of Arl1 and ARF1. arfaptin2 or arfaptin1 was transformed into yeast AMR70 and mate with yeast strain L40 transformed with one of the small GTPase as indicated. Beta-gal assays were performed to dissect the interactions.
28
Fig 14. Arl1QLd17N decreases the Golgi association of arfaptin2. Cos-b cells were transiently transfected with constructs expressing Arl1wt or Arl1QL or Arl1QLd17N or Arl1TN. Cells were fixed and double labeled with a antibody against endogenous arfaptin2 and a anti-myc antibody to reveal the C terminal myc tagged Arl1. Arl1QL without the N terminal 17 residues evenly
29
Fig 15. Arl1QL interacts with ApoC2. ApoC2 or ApoB were transformed into yeast AMR70 respectively and mate with yeast strain L40 transformed with one of the small GTPase as indicated. Beta-gal assays were performed to dissect the interactions.
30
Fig 16. ARFGAP1 changes the patterns of Golgi proteins such as Arl1, golgin245 and golgin97, but ARFGAP1 can’t diffuse the overexpressed ARF1QL. Cos-b cells were transiently transfected with constructs expressing GFP-tagged ARFGAP1 or cotransfected with ARF1QL-myc as indicated. Cells were fixed and double labeled with antibodies against Arl1 or golgin245 or golgin97 or a antibody against myc tag to detect myc-tagged ARF1QL.
31
Fig. 17. Analysis of Arl1 protein complex based on affinity purification method coupling with two-dimensional electrophoresis. The Arl1QL gene was cloned to pcDNA-TAP plasmid as a C-terminal tagged fusion protein. After 48 h transfected to 293 T cells, cells were harvested, lysed and then incubated with IgG Sepharose beads. The protein complex was purified by elution with 0.5 M acetic acid (pH 3.4) following trichroroacetic acid precipitation. Both protein complexes purified from Arl1Ql-TAP and TAP constructs were applied to an immobilizes pH gradient gel strip containing pH range of 3-10 ampholytes, and isoelectric focusing was performed. After isoeletric focusing, the strip was subjected on 12.5% gels visualized by silver stained.
1 2
TAP hARL1QL-TAP
A. B.
32
4. Projected Timeline & Brief Summary of Plans for Next Year
There are still many gaps in our understanding of roles of specific ARL proteins in nuclear dynamics. As several mammalian ARFs and ARLs have been described, it seemed reasonable that they should have different functions. The ARL1 provides an opportunity to define their roles in another kind of membrane trafficking and/or nuclear dynamics. The results from our proposed work should also provide important contribution on the understanding of this trafficking mechanism. In particular, we are very likely to have the following contributions.
First, we will identify several regulators or effectors that interact specifically with ARL1, and further study their activity in nuclear dynamics. We should identify those components, which are necessary for optimal stimulus-effector coupling for ARL activity. If the separation of proteins is not good enough, two-dimension gel separation will be carried out. Antibodies against proteins identified by affinity-purification will be generated. These antibodies will be used to confirm their specific interaction with ARL1 by pulling down the immunoprecipitated complexes in vivo. Using immunoprecipitaiton, we may identify different ARL1-complexes, which have other auxiliary factors at different physiological conditions. Using yeast genetics can give us a quick analysis of the existing factors. In addition, we will also use chemical cross-linking techniques and immuno-precipitation assay to determine their specific interaction. The antibodies against recombinant ARL1 interacting proteins will be used to identify their subcellular localization. We will further use these antibodies to study their cellular interaction. Our results will show how ARLs interact specifically with GEF and GAP and how these interactions execute nuclear dynamics signaling.
Second, subcellular localization of ARL1, and their mutated forms will be determined. Substrate and lipids specificity of these purified proteins will be characterized. Effects on translocation of specific-marker proteins in stable transfectants, expressing dominant negative mutants. This will also clarify whether ARL1 have similar regulatory activity. Thus, we may have some clues on the functional roles of ARL1 in Golgi to lysosome trafficking.
Third, we will demonstrate that ARL1 and its interacting proteins can shuttle between trans-Golgi and lysosome. We will also identify hARL1-complex proteins and their specific transport cargos. We will determine the hARL1-complex in Golgi-lysosome sorting function. In addition, we will determine sorting signal and binding partners of the ARL1 regulated transport vesicles.
This project offers an excellent chance to train graduate students and postdoctoral fellows. Especially, the proposal integrates many disciplines including molecular biology, biochemistry, genetics, and cell biology. Not only the students will get in-depth training but also will obtain a broad view and understand the interaction of these disciplines.
33 5. Personnel
Summarize the personnel involved in the project during the grant period. List the personnel in accordance to the following categories: (1) senior investigators, including visitors; (2) postdoctoral fellows; (3) graduate students; (4) technicians or research assistants. Specify for each individual the period of involvement and the percentage commitment of effort.
Name In Chinese In English Position Title Education Degree % of personal effort on this project Job Description or Responsibilities 許嘉伊 吳宗聖 李綺華 卓逸明 graduate student graduate student graduate student research assistant MS BS MS MS 100% 100﹪ 70% 100% Functional characterization of ARLs and their interacting molecules
Cloning and expression of ARLs gene
Functional characterization of ARLs and their interacting molecules
Cloning, expression, and localization of ARLs and their mutants.
34 6. Publications and/or Patents
6a. Publications
List the title and complete references (author(s), journal or book, year, page number) of all publications directly resulting from studies supported by the project (i.e., with citation of this grant in the acknowledgement section). List the publications for the project in accordance to the following categories: (1) manuscripts published and accepted for publications; (2) manuscripts submitted; and (3) conference proceedings. Provide one copy of each publication not previously reported to the National Science Council in the Appendix.
35 6b. Patents
List all inventions disclosed, patents filed, and patents granted. Please note the inventors, assignee, title of patent, country or area where patent applied for, filing or issued number and date.
36