國立交通大學
奈米科技研究所
碩
士
論
文
超順磁奈米粒子之強磁場誘發線蟲之細胞自然凋亡並影響其行為
Induction of apoptosis and reduced mobility for Caenorhabditis elegans by
super-paramagnetic nanoparticles enhanced magnetic field
研 究 生:葉 禮 閣
學 號:9452520
指導教授:黃 國 華 副教授
超順磁奈米粒子之強磁場誘發線蟲之細胞自然凋亡並影響其行為
學生:葉禮閣 指導教授:黃國華 副教授國立交通大學奈米科技研究所 碩士班
中文摘要
在現代生活中,我們週遭都有磁場,而且是不可避免的。一般的家電用品、手機、 醫療的 NMR 等,都會產生磁場,且磁場是超距力,說我們的生活在磁場底下真的ㄧ點 也不為過。就我們的經驗,我們知道磁場可能會對生物造成不良的影響,但ㄧ直缺乏科 學上的證據,在過去 20 幾年,科學家不斷希望找出磁場對生物的影響,但到目前為止, 仍然沒有找到直接的科學證據。 現在我們利用線蟲(C.elegans)這種簡單的模式生物,用簡單的強磁裝置加上超順磁 奈米粒子,我們模擬生物在強磁下的環境,觀察行為與基因表現的差異,找出磁場對生 物影響的生理因子,我們發現了磁場會導致自然凋亡(apoptosis),並提出基因表現的證 據。 關鍵字: 線蟲、磁場、自然凋亡Induction of apoptosis and reduced mobility for Caenorhabditis elegans by super-paramagnetic nanoparticles enhanced magnetic field
Student: Li-Ko Yeh Advisors: Dr. Guewha Steven Huang
Institute of Nanotechnology National Chiao Tung University
Abstract
Intuitively, the environmental electro-magnetic field may cause bad influence and bring abnormalities to life, such as cancer. For decades, scientist worked hard to look for the association between elctro-magnetic field and diseases desperately. The direct evidence, however, is yet to be identified.
In the current study, we employed Caenorhabditis elegans as a model and applied localized strong magnetic field through administration of super-paramagnetic nanoparticles fed into the worms.
We found that the 3-day treatment of magnetic field caused significant retardation to the mobility of C. elegans. A reduction of 50 % in mobility was observed to the N2 L4 worm treated with localized magnetic field for 3 days. To investigate possible molecular mechanisms underlying the retardation of mobility, real-time reverse transcriptase polymerase chain reaction (RT-RT PCR) was performed using more than 100 pairs of primer specific to genes directly associated with apoptosis, oncogenesis, and oxidative stress. Expression levels were altered for at least 8 folds for more than 50 genes tested in this study.
To validate the correlation between magnetic field and apoptotic genes, ced-3, ced-6, and cbp-1mutant worms were obtained from CGC and were tested for their sensitivity to the magnetic field. Ced-3 mutant worm was insensitive to magnetic field, while ced-6 and cbp-1 mutant worms were sensitive to magnetic field. The lost of sensitivity to magnetic field in the ced-3 mutant strongly suggested that apoptosis was caused by the magnetic field and since
ced-3 is lack of apoptosis thus is insensitive to the magnetic field. Sensitivity of ced-6 and cbp-1mutants was expected due to their assistant role in the apoptosis.
In conclusion, the current study provided direct evidences that magnetic field caused apoptosis which led to the retardation for the mobility.
誌 謝
首先誠摯的感謝指導教授黃國華博士及師母,人非生而知之者,孰能無惑。 老師悉心的教導使我得以與陌生 C.elegans 領域的結緣,時時刻刻的討論並指點我正確 的方向,使我在兩年中獲益匪淺。老師對學問的嚴謹更是我輩學習的典範。師母為人正 氣凜然,有ㄧ代俠女之風,每日的噓寒問暖更使我感受在心。 本論文的完成另外亦得感謝的洪耀欽教授不少的建議。因為有長輩們的體諒及幫 忙,使得本論文能夠更完整而嚴謹。 山不在高,有僊則名。水不在深,有龍則靈,我們的「陋室」,有「老師與師母」 則行。兩年裡的日子,實驗室裡共同的生活點滴,實驗室從一開始人家不要廢墟到現在 充滿學術氣息的實驗室,裡面的每個部分都是我珍貴的回憶,實驗室給我許多磨練與成 長的機會,除了學術上的討論、做人與做事的態度外、更感謝眾位學長姐、同學、學弟 妹的共同努力,你/妳們的陪伴讓兩年的研究生活變得絢麗多彩。 感謝張凱明、汪孟德、陳昱勳學長、不厭其煩的指出我研究中的缺失,且總能在我 迷惘時為我解惑,也感謝蘇嘉緯、陳永昌、林志杰同學的幫忙,恭喜我們順利走過這兩 年。「如果說我看的比別人更遠,那是因為我站在巨人的肩膀上。」因為我站在各位的 肩膀上,不讓狹隘的目光和偏見,遮蔽了自己的眼界,讓自己喪失欣賞美麗風景的機會, 在次感謝這兩年研究生捱中的每個巨人,使我的視野才能更加遼闊。Contents
Chapter 1 Introduction
1.1 Does the magnetic field has influence on the life---1
1.1.1 Magnetic field past research---1
1.1.2 Magnetic field currently research---2
1.2 Magnetic field influence with inadequate evidence---2
1.2.1 Magnetic field influence in animal---3
1.3 Other research in magnetic field---3
1.4 Caenorhabditis elegans (C.elegans)---4
1.4.1Caenorhabditis elegans V.S. magnetic field---5
1.4.2 C.elegans -a kind of very good research source material---5
1.5 Nanoparticles---6
1.5.1 Fe3O4 is nature magnet---6
1.5.2 TiO2 , Fe2O3 & Fe3O4 toxicity ---6
1.6 Magnetic field & Gene expression---7
1.7 Apoptosis & gene ---7
1.7.1 Human apoptosis related gene &apoptosis pathway---8
1.7.2 C.elegans apoptosis & cancer related gene in wormbase---9
Chapter 2 Research experiment & design---10
2.1 C.elegans culture---10
2.2 Magnetic field design---10
2.3 Video record system---12
2.4 Experiment flow chart---12
2.4.1 Flow chart of gene analysis---13
2.5 SDS-PAGE GEL---13
2.5.1 SDS-PAGE protocol---14
2.6 Primer design---16
2.6.1 Gene primer designed---16
2.6.2 Gene primer designed list---17
2.7 RNA Concentration and Quality---20
2.7.1 RNA extraction protocol---20
2.8 Reverse Transcriptase from RNA to cDNA---22
2.8.1 Reverse Transcriptase protocol---23
2.9 Polymerase chain reaction (PCR) principle---24
2.9.1 PCR protocol---25
2.10 Real Time – PCR principle---25
2 C 3.1 3. 3. 3. 3. 3.5 3.7 3 3. 3. hapter 5 Reference---63
2.11.1 DNA gel protocol---28
hapr 3 Result & Discussion---30
C.elegans behavior---30
3.1.1 Video recorded---31
2 Super-paramagnetic nanoparticles in C.elegans---37
3.2.1Super-paramagnetic nanoparticles-Fe3O4---37
3.2.2 Optics microscope picture---37
3.3 C.elegans SDS-page---40 4 PCR data---41 3.4.1 Gene number---41 3.7.1 .PCR electrophoresis---41 5 Real-time PCR data---45 5.1 Real-time PCR system---45
3.5.1.1 Real –time PCR procedure---45
3.5.2 Gene experission---46
3.5.3 Fist time real-time PCR test---46
.4 Real-time PCR data believable---48
3.5.4.1 Ced family gene expression---49
3.5.4.2 Other apoptosis related gene expression---51
3.5.4. Some cancer related gene expression---51
3.6 C.elegans GFP marked---52
C.elegans mutant behavior & real-time PCR data---53
.7.1 Ced-3 MT1522---55
3.7.1.1 Ced-3 mutant behavior---55
3.7.1.2 Ced-3 mutant real-time PCR data---56
7.2 Ced-6 MT4970---57
3.7.2.1 Ced-6 mutant behavior---57
3.7.2.2 Ced-6 mutant real-time data--- 58
7.3 Cbp-1 VC1006---59
3.7.2.1 Cbp-1 mutant behavior---59
3.7.2.2 Cbp-1 mutant real-time data---60
Chapter 4 Conclusion---61 C
3
Figure Contents
Fig 1 C.elegans anatomy---4
Fig 2.Apoptosis pathway---8
Fig 3. Wormbase website---9
Fig 4 .Magnetic field for experiment---10
Fig 5. Manufacturer's information---11
Fig 6.Video record system---12
Fig 7. Experiment flow chart---12
Fig 8.Flow chart of gene analysis chart---13
Fig 9.SDS-PAGE device & sample---13
Fig 10.Gene primer classification---16
Fig 11 RNA quality gel---21
Fig 12. mRNA to cDNA---22
Fig 13.PCR principle---24
Fig 14. Real-time PCR principle---26
Fig 15.Real-time PCR CT analyse principle---27
Fig 16. SYBR Green formula---27
Fig 17. Electrophoresis principle---29
Fig 18. C.elegans behavior with mannetic field---30
Fig 19. Control-N2 mobility in succession---32
Fig 20. N2+Fe3O4 mobility in succession---33
Fig 21. N2+magnetic field mobility in succession---34
Fig 22. N2+Fe3O4+magnetic field 48hr mobility in succession---35
Fig 23. N2+Fe3O4+magnetic field 96hr mobility in succession---36
Fig 24. N2 optics microscope picture---38
Fig 25. Fe3O4 in C.elegans alimentary canal---39
Fig 26. C.elegans SDS-page---40
Fig 27.Gene PCR gel 1-25---42
Fig 28.Gene PCR gel 26-50---42
Fig 29.Gene PCR gel 51-74---43
Fig 30.Gene PCR gel 75-99---43
Fig 31.Gene PCR gel 100-109---44
Fig 32. Repeat gene PCR gel 76-96,L18,L21---44
Fig 33. Real –time PCR procedure---45
Fig 34. Fist time real-time PCR data test---47
4
Fig 38.Other apoptosis related gene real-time PCR data to gene expression---50
Fig 39.Some cancer related gene real-time PCR data to gene expression---51
Fig 40. GFP mark signal---52
Fig 41.GFP signal decrease with magnetic field---52
Fig 42 Ced-3,ced-6,cbp-1 mutant location genomic environs---54
Fig 43. Ced-3 mutant behavior with magnetic field---55
Fig 44. Ced-3 mutant real-time PCR data to gene expression---56
Fig 45. Ced-6 mutant behavior with magnetic field---57
Fig 46. Ced-6 mutant real-time PCR data to gene expression---58
Fig 47.Ced-6 apoptosis related pathway---58
Fig 48. Ced-6 mutant behavior with magnetic field---59
Fig 49. Cbp-1 mutant real-time PCR data to gene expression---60
Fig 50 Reference [33] some data---62
Table Contents
Table1. Gene primer designed list---20Table 2. C.elegans behavior Classification---30
Table.3 Gene number list---41
Chapter 1
Introduction
1.1 Does the magnetic field have influence on the life?
EMF is everywhere and can not be avoiding until daily life. It is magnetic fields to the influence on the biological behavior or the mechanism that we care about the focus of this fact sheet is on extremely low-frequency magnetic fields. Examples of devices that emit these fields include power lines and electrical appliances, such as electric shavers, hair dryers, computers, televisions, electric blankets, NMR, and heated waterbeds. Most electrical appliances have to be turned on to produce a magnetic field. The strength of a magnetic field decreases rapidly with increased distance from the source. Of course, as more and more magnetic fields of the development in science and technology are suitable for our daily life.
There is no conclusive evidence that EMF may have effect. We believe that this is violating with the intuition; there is an enormous body of evidence later, but science is to stress the evidence. The evidence seems to be very poor. Therefore, we will be presenting a list of papers and ORs which either show serious effects or are considered important papers on the subject which we have collected over the years.
The progress of nanotechnology helps us to research this topic. We utilize Fe3O4 - ultra paramagnetic nanoparticles, and the gene expression to prove magnetic field impacting on life.
1.1.1
Magnetic field past research
In the past research, numerous epidemiological (population) studies and comprehensive reviews have evaluated magnetic field exposure and risk of cancer in people. Epidemiologic and experimental research on the potential carcinogenic effects of extremely low frequency electromagnetic fields (EMF) has now been conducted for over two decades. Cancer epidemiology studies in relation to EMF have focused primarily on brain cancer and leukemia, both from residential sources of exposure in children and adults and from occupational exposure in adult men [1]. No consistent relationship has been seen in studies of children brain tumors or cancers at other sites and residential ELF electric and magnetic fields [2].
However, these studies have generally been smaller and of lower quality Four long-term bioassays have been published in which the potential oncogenicity in experimental animals of exposure to ELF magnetic fields was evaluated in over 40 different tissues using standard chronic toxicity testing designs [1, 2]. Since the two most common cancers in children are leukemia and
A study in 1979 pointed to a possible association between living near electric power lines and childhood leukemia .The finding was strongest for childrenwho had spent their entire lives at the same address, and itappeared to be dose-related. It did not seem to be an artifactof neighborhood, street congestion, social class, or familystructure. The reason for the correlation is uncertain; possibleeffects of current in the water pipes or of AC magnetic fieldsare suggested. [3].
1.1.2 Magnetic field currently research
Among more recent studies, findings have been mixed. Some have found an association; others have not. These studies are discussed in the following paragraphs. Currently, researchers conclude that there is limited evidence that magnetic fields cause childhood leukemia, and that there is inadequate evidence that these magnetic fields cause other cancers in children [2]. Researchers have not found a consistent relationship between magnetic fields or appliances and childhood brain tumors.
1.2 Magnetic field influence with inadequate evidence
Is it harmful not to have direct evidence at present to children? However , before research let me know the magnetic field has suspicion to the danger of the health. For adults, a Norwegian study found a risk for exposure to magnetic fields in the home [4], and a study in African-American women found that use of electric bedding devices may increase breast cancer risk [5].Some occupational studies showed very small increases in risk for leukemia and brain cancer, but these results were based on job titles and not actual measurements. More recently conducted studies that have included both job titles and individual exposure measurements have no consistent finding of an increasing risk of leukemia, brain tumors, or female breast cancer with increasing exposure to magnetic fields at work .Cases of breast cancer diagnosed during 1980–1996were identified in a cohort of women living near a high-voltage power line in Norway in 1980 or between 1986 and 1996. Women with the highest occupational exposure had an odds ratio of 1.13 (95% CI: 0.91, 1.40) when compared with those unexposed at work [4].
The findings suggest an association between exposure to magnetic fields and breast cancer in women. A total number of 608 cases and 667 controls participated. Adjusting for accepted breast cancer risk factors, we found an OR of 1.13 for lifetime occupational exposure to ELF-MF at medium or high intensities [6]. Risks were larger for exposures before age 35 (OR¼1.40), and statistically significant for exposures before 35 among cases with progesterone receptor positive tumors (OR¼1.56, 95% CI¼1.02–2.39).In a word, there appears to be a small increased risk for breast cancer among postmenopausal women exposed occupationally to ELF-MF. [7, 8]
1.2.1
Magnetic field influence in animal
Speaking of the influence that the magnetic field endangers to the health, does the magnetic field to other living thing work? In order to distinguish this, scientists learned from animal experiments about the relationship between magnetic field exposure and cancer. Studies to evaluate immune function and host resistance in animals have given negative effects for exposure to ELF electric and magnetic fields (2). In-vitro exposure of immune system cells generally did not because changes in proliferation capacity .The few animal studies on cancer-related non-genetic effects are inconclusive. Results on the effects on invitro cell proliferation and malignant transformation are inconsistent, but some studies suggest that ELF magnetic fields affect cell proliferation and modify cellular responses to other factors such as melatonin. An increase in apoptosis following exposure of various cell lines to ELF electric and magnetic fields has been reported in several studies with different exposure conditions. Numerous studies have investigated effects of ELF magnetic fields on cellular end-points associated with signal transduction, but the results are not consistent (2). The absence of animal data supporting carcinogenicity makes it biologically less likely that magnetic field exposures in humans, at home or at work, are linked to increased cancer risk [17].
1.3 Other research in magnetic field
Impact on nervous system: Some research indicate low magnetic field make the active increase of the animal often, excitability increases; And stronger magnetic field often makes the activity of the organism reduce, excitability is reduced, suppress reacting. [14]
In addition, the magnetic field also has certain function on the plant nerve, to the heartbeat, blood pressure; breathe certain influence .Impact on cardiovascular system: The magnetic field does not have obvious function on the heart under the normal condition, but to the heart under the pathology state, play certain treatment to act on. Impact on immunity[15]., internal system: Because of the difference in method, means, condition of studying,etc., have disputes to the function on the immune system of the magnetic field at present. But most researchers think, the magnetic field can improve the immune function of the organism. The magnetic field still has a secretion activity of regulating endocrine glands; strengthen the secretion of the active material of immunity.
1.4 Caenorhabditis elegans (C.elegans)
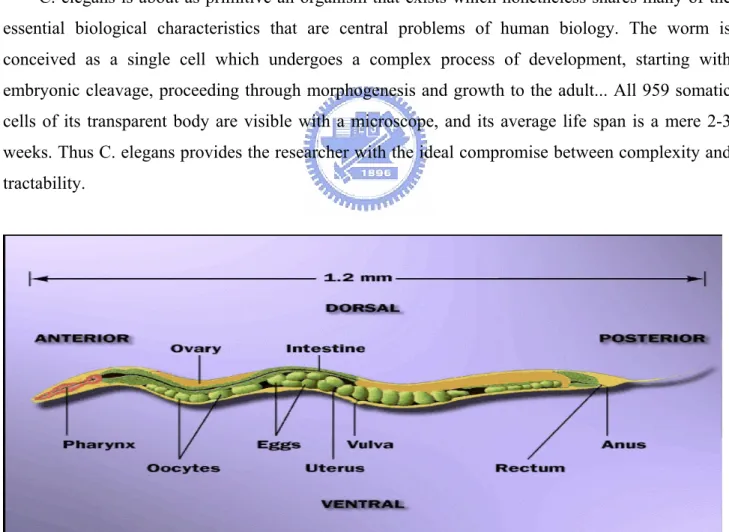
Caenorhabditis elegans(Fig 1) is a nematode - a member of the phylum Nematoda: “Nematoda. The roundworms and threadworms, a phylum of smooth-skinned, unsegmented worms with a long cylindrical body shape tapered at the ends; includes free-living and parasitic forms both aquatic and terrestrial” It is small, growing to about 1 mm in length, and lives in the soil - especially rotting vegetation - in many parts of the world, where it survives by feeding on microbes such as bacteria. It is of no economic importance to man. (Academic press Dictionary of Science and Technology) Around the world many hundreds of scientists are working full time investigating the biology of C. elegans. Between October, 1994 and January, 1995\ 73 scientific articles about this creature appeared in international science journals. Currently an international consortium of laboratories are collaborating on a project to sequence the entire 100,000,000 bases of DNA of the C. elegans genome.
C. elegans is about as primitive an organism that exists which nonetheless shares many of the essential biological characteristics that are central problems of human biology. The worm is conceived as a single cell which undergoes a complex process of development, starting with embryonic cleavage, proceeding through morphogenesis and growth to the adult... All 959 somatic cells of its transparent body are visible with a microscope, and its average life span is a mere 2-3 weeks. Thus C. elegans provides the researcher with the ideal compromise between complexity and tractability.
1.4.1Caenorhabditis elegans V.S. magnetic field
Change of the magnetic field, the biological behavior of the Caenorhabditis elegans is influenced. For one thing, high magnetic fields on biological process, by which alternating current magnetic stimuli as high as 1.7 T can be administered. Available evidence indicates that alternating high magnetic fields can elicit both chronic and acute biological effects but that the effects may be well tolerated or compensated for by the living organism [9]. Transgenic nematodes (Caenorhabditis elegans strain PC72), carrying a stressinducible reporter gene (escherichia coli betagalactosidase) under the control of a C. elegans hsp16 heatshock promoter, have been used to monitor toxicant responses both in water and soil[10]. For another, effect of extremely low frequency electromagnetic fields (ELFEMF) in the presence of a second stressor (mild heat shock) on the expression of a lacZ reporter gene under the control of hsp16 or hsp70 promoters in two transgenic strains of C. elegans. [11].Exposure of Caenorhabditis elegans to extremely magnetic fields induces stress responses. Expression of the hsp16lacZ gene was enhanced when transgenic animals were exposed to magnetic fields up to 0.5 T at 60 Hz. The hsp promoter was more efficiently expressed at the embryonic than at the postembryonic stage irrespective of exposure.Caenorhabditis elegans, revealed that intermittent exposure to the magnetic fields modestly inhibited the animal's reproduction as well as its post-embryonic development. [12]
1.4.2 C.elegans -a kind of very good research source material
C. elegans is a free-living nematode. There are two sexes: a self-fertilizing hermaphrodite and a male. The adult essentially comprises a tube, the exterior cuticle, containing two smaller tubes, the pharynx and gut, and the reproductive system. Most of the volume of the animal is taken up by the reproductive system. Of the 959 somatic cells of the hermaphrodite some 300 are neurons. Neural structures include a battery of sense organs in the head which mediate responses to taste, smell, temperature and touch - and although C. elegans has no eyes, it might respond slightly to light. Among other neural structures is an anterior nerve ring with a ventral nerve cord running back down the body. (There is also a smaller dorsal nerve cord.) There are 81 muscle cells. C. elegans moves by means of four longitudinal bands of muscle paired sub-dorsally and sub-ventrally. Alternative flexing and relaxation generates dorsal-ventral waves along the body, propelling the animal along. The development and function of this diploid organism is encoded by an estimated 17,800 distinct genes.
http://www.wormbase.org/ http://www.cbs.umn.edu/CGC/ http://www.celeganskoconsortium.omrf.org http://www.wormatlas.org/handbook/contents.htm
Biological effect of the magnetic field: The magnetic field is influential to the organism, include the following aspects mainly:
1.5 Nanoparticles
In our experiment, we use Fe3O4 (6-10nm), Fe2O3 (30nm) and TiO2 (10-40nm).
1.5.1 Fe3O4 is nature magnet
It likes nature little magnet in the body. Most materials found in organisms are generally thought of as being nonmagnetic- for example, C.elegans[13],either diamagnetic (repelled weakly from a magnetic field, as is water and almost any fatty substance) or paramagnetic (weakly attracted to a magnetic field, as is deoxyhemoglobin in blood cells [21].
1.5.2 TiO2, Fe2O3 and Fe3O4 toxicity
Fe3O4, Fe2O3, and TiO2 had no measurable effect on the cells toxicity. Toxicity of iron oxides and metabolites of benzo apyrene alone or in combination in cells culture and identification by laser microprobe mass spectrometry.[22] These results suggest that Fe2O3 and Fe3O4 alone are not very toxic but the association of one of these compounds with BaP increases the toxicity of the latter. On the other hand, TOF-LMMS seems to show a metabolization of iron oxide into reduced form. But, it is necessary to raise the ambiguity about the iron which is always in the cells. \TiO2 had no measurable effect on the cells until the concentrations reached greater than 200 microg/mL [23]. LDH leakage significantly increased in the cells exposed to ZnO (50 to 100 microg/mL), while other nanoparticles tested displayed LDH leakage only at higher doses (>200 microg/mL).
1.6 Magnetic field and Gene expression
Exposure to magnetic field (5 mT at 60 Hz) does not affect cell growth and c-myc gene expression designed and manufactured equipments for long-term and low-density (0 to 9 mT) exposures of cultured cells to extremely low frequency magnetic fields (ELF-MF), and examined the effects of ELF-MF on cell growth and c-myc mRNA expression in Chinese hamster ovary (CHO) cells [16].
The wide-spread induction of stress-related genes and transcription factors, and a depression of genes associated with cell wall metabolism, are prominent examples. Magnetic Field-Responsive Sequences in hsp70. [18] HSP70 gene expression is induced by a wide range of environmental stimuli, including 60-Hz electromagnetic fields. In an earlier report we showed that the induction of HSP70 gene expression by magnetic fields is effected at the level of transcription and is mediated through c-myc protein binding at two nCTCTn sequences at 2230 and 2160. In the human HSP70 promoter. report on the identification of a third c-myc binding site (between 2158 and 2162) that is an important regulator of magnetic field-induced HSP70 expression.We also show that the heat shock element (HSE), lying between 2180 and 2203, is required for induction of HSP70 gene expression by magnetic fields[20]. High magnetic field induced changes of gene expression in
Arabidopsis[19) Field strengths in excess of about 15 Tesla induce expression of the Adh/GUS transgene in the roots and leaves. From the microarray analyses that surveyed 8000 genes, 114 genes were differentially expressed to a degree greater than 2.5 fold over the control. These results werequantitatively corroborated by qRT-PCR examination of 4 of the 114 genes [19].
1.7
Apoptosis and gene
Apoptosis is mediated by a family of cysteine-aspases (caspases), which are expressed as inactive zymogens and are prototypically processed to an active state following an apoptotic stimulus. Many important human diseases are caused by abnormalities in the control of cellular apoptosis (programmed cell death). These abnormalities can result in either a pathological increase in cell number (e.g. cancer) or a damaging loss of cells (e.g. degenerative diseases).
1.7.1 Human apoptosis related gene and apoptosis pathway
The intrinsic pathway (Fig 2) requires disruption of the mitochondrial membrane and the release of mitochondrial proteins including Smac/DIABLO, HtRA2, and cytochrome c. Cytochrome c functions with Apaf-1 to induce activation of caspase-9, thereby initiating the apoptotic caspase cascade, while Smac/DIABLO and HtrA2 bind to and antagonize IAPs. Mitochondrial membrane permeabilization is regulated by the opposing actions of pro- and antiapoptotic Bcl-2 family members. Multidomain proapoptotic Bcl-2 proteins (e.g., Bak and Bax) can be activated directly following interaction with the BH3-only Bcl-2 protein Bid. Alternatively, binding of other BH3-only proteins (e.g., Noxa, Puma, Bad, and Bim) to antiapoptotic Bcl-2 proteins (e.g., Bcl-2 and Bcl-XL) results in activation of Bax and Bak. The regulated release of proapoptotic factors from the mitochondria cause induction of downstream caspases, and potential loss of mitochondrial function [28].
Fig 2.Apoptosis pathway
Cells have a discrete cell death pathway defined by a specific set of genes. These genes encode proteins that form the biochemical cascade that ultimately leads to cell death (Fig. 2). The key genes that control the cell death process are the cell death effectors of the CED-3/ICE ("caspase") family and the cell death inhibitors of the BCL2 family [29].
We suppose the magnetic field induction to apoptosis.
1.7.2 C.elegans apoptosis and cancer related gene in wormbase
Form wormbase website (Fig 3), we can find thousands of kinds of genes about the C.elegans. Among which are about apoptosis, cancer and nerve, etc.
How could we use wormbase? Wormbase is database of an introduction C.elegans, include data of nematode genome inside, main function
(1) sequencing, genome of C.elegans has already been solved out, can find the materials of the array on this website.
(2) Mapping can find out the gene location is in C. elegans which of one chromosome at the position (3) Phenotypic information has database which lists RNAi to C. elegans
If you want to experiment as RNAi of model organism, you can consult this website.I find some genes, as apoptosis related gene, unusual function with magnetic field in C. elegans
Fig 3. Wormbase website
Chapter 2
Research experiment and design
2.1 C.elegans culture
The nematode Caenorhabditis elegans has proven to be an important model organism for. aging-related research, providing a well characterized paradigm for NGM + 50 μM FUDR (DR-FD media) culture at 20oC
2.2 Magnetic field design
Fig 4 . Magnetic field for experiment
What we work is research that under the function of the magnetic field C.elegans biological behavior and physiological mechanism enable denying changing?
Yes .Change in our experiment.
We have designed a series of experiments and inferred for this the gene expression.
Fig 5. Manufacturer's information
Force the nematode to grow under stabilizing and covering on the function of the magnetic field with my design of the experiment. We use 6cm plate and cover and invite 3mm agar. Because the magnetic field is inversely proportional to the square of distance strongly Make use of this best magnetic field which we can obtain of method (Fig 4) .Our magnetic field that quantity examine is 0.5T on.
2.3 Video record system
We combines against Sony video cassette recorder three-dimensionally and microscope notebook computer (Fig 6). This kind of method lowers costs, and the result is very good too. In me experiment, I 8design many device that not been used before this.
Fig 6.Video record system
2.4 Experiment flow chart
Fig 7. Experiment flow chart
Magnetic design
Behavior different biochemical action
protein SDS-page Apoptosis Cancer Oxidation Related gene MAX difference Normal condiction RT RT-PCR EMF V.S.change NO YES Gene express
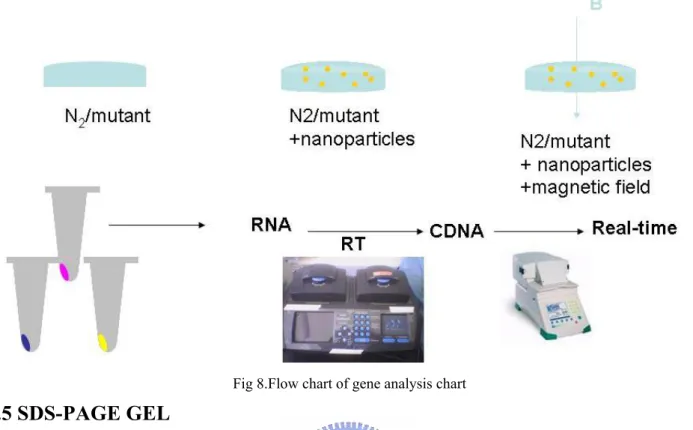
2.4.1Flow chart of gene analysis
Fig 8.Flow chart of gene analysis chart
2.5 SDS-PAGE GEL
SDS-PAGE、officially sodium dodecyl sulfate polyacrylamide gel electrophoresis、is a technique used in biochemistry、genetics and molecular biology to separate proteins according to their electrophoresis mobility (a function of length of polypeptide chain or molecular weight as well as higher order protein folding、posttranslational modifications and other factors).
2.5.1 SDS-PAGE protocol
Materials
30% acrylamide 10% SDS
10% APS (make fresh each time) TEMED
1.5 M Tris, pH 8.8 (resolving gel) 1.0 M Tris, pH 6.8 (stacking gel) 5x SDS Running Buffer (1 L) Tris 15 g
Glycine 72 g SDS 5 g
Coomassie Blue Stain 10% (v/v) acetic acid
0.006% (w/v) Coomassie Blue dye 90% ddH2O
Isopropanol Fixing Solution 10% (v/v) acetic acid 25% (v/v) isopropanol 65% ddH2O
SDS sample loading buffer (40 ml) ddH2O 16 ml
0.5 M Tris, pH 6.8 5 ml 50% Glycerol 8 ml 10% SDS 8 ml
2- mercaptoethanol 2 ml (add immediately before use) bromophenol blue
10% (v/v) acetic acid Protocol
1. Prepare polyacrylamide gel according to standard protocol.
2. Load samples and run gel @ 25 mA (2 gels run @ 50 mA) in 1x SDS Running Buffer.
3. At this point, the gel can either be transferred to a membrane (see Western protocol) or stained with Coomassie (see below).
4. Place gel in a plastic container. Cover with isopropanol fixing solution and shake at room temperature. For 0.75 mm-thick gels, shake 10 to 15 min; for 1.5 mmthick gels, shake 30 to 60 min.
5. Pour off fixing solution. Cover with Coomassie blue staining solution and shake at RT for 2 hr.
6. Pour off staining solution. Wash gel with 10% acetic acid to destain, shaking at RT ON.
2.6 Primer design
The gene of interest usually has to be amplified from genomic or vector DNA by PCR (polymerase chain reaction) before it can be cloned into an expression vector. The first step is the design of the necessary primers.
Important features are:
Primer sequence. Especially the 3'-end of the primer molecule is critical for the specificity and sensitivity of PCR. It is recommemded not to have:
Primer pairs should be checked for complementarity at the 3'-end. This often leads to primer-dimer formation . Bases at the 5'-end of the primer are less critical for primer annealing. Therefore, it is possible to add sequence elements, like restriction sites, to the 5'-end of the primer molecule.
We use a primer length of 18-23 bases is optimal for our PCR applications Find out the gene that we are interested in from wormbase . Make using of knowning gene sequence to design the primers. ..
2.6.1 Gene primer designed
We look for the suitable gene correlated with our design environment as much as possible
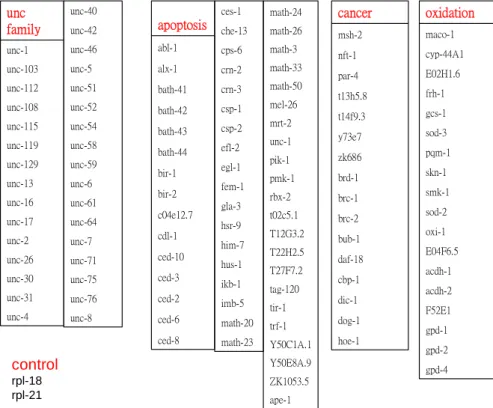
With the behavior and observation of C.elegans, we roughly sum up into the gene that four groups are correlated with and form a cluster.Among them it is UNC family correlated with the behavior , cancer, apoptosis and anti-oxidant groups(Fig 10). Verified in the past research that there are a large amount of databases for guessing in the the genome
gpd-4 gpd-2 gpd-1 F52E1 acdh-2 acdh-1 E04F6.5 oxi-1 sod-2 smk-1 skn-1 pqm-1 sod-3 gcs-1 frh-1 E02H1.6 cyp-44A1 maco-1 oxidation hoe-1 dog-1 dic-1 cbp-1 daf-18 bub-1 brc-2 brc-1 brd-1 zk686 y73e7 t14f9.3 t13h5.8 par-4 nft-1 msh-2 cancer ced-8 ced-6 ced-2 ced-3 ced-10 cdl-1 c04e12.7 bir-2 bir-1 bath-44 bath-43 bath-42 bath-41 alx-1 abl-1 apoptosis math-23 math-20 imb-5 ikb-1 hus-1 him-7 hsr-9 gla-3 fem-1 egl-1 efl-2 csp-2 csp-1 crn-3 crn-2 cps-6 che-13 ces-1 ape-1 ZK1053.5 Y50E8A.9 Y50C1A.1 trf-1 tir-1 tag-120 T27F7.2 T22H2.5 T12G3.2 t02c5.1 rbx-2 pmk-1 pik-1 unc-1 mrt-2 mel-26 math-50 math-33 math-3 math-26 math-24 unc-4 unc-31 unc-30 unc-26 unc-2 unc-17 unc-16 unc-13 unc-129 unc-119 unc-115 unc-108 unc-112 unc-103 unc-1 unc family unc-8 unc-76 unc-75 unc-71 unc-7 unc-64 unc-61 unc-6 unc-59 unc-58 unc-54 unc-52 unc-51 unc-5 unc-46 unc-42 unc-40 control rpl-18 rpl-21
2.6.2 Gene primer designed list
Fwd sequence Rev sequence
unc-1 TAGAACGTGTGGAAGTGAAGG GTTGCAAATCGGAATTTGG unc-103 CCAAGCAACTTGCACAACC GTCGTTCGAAACCTCATCG unc-112 GAAGAAGCCACACTATTTGC CACCTCCTGTTGATATGACG unc-108 GGAAAATCCTGCTTGCTCC CTCATTGTTAATGTCGAACACG unc-115 ACGGGCTTCTACACAACACC CACACTCGTAACATTTGACACC unc-119 CTTCCAGGAATCACTCAAGG GGGAAATTATGCATCATACG unc-129 CGACTTCCGATTGTCTTATTGC CTTCCAAATTCTCATCTCCTCC unc-13 CCATTAAAGACAGTTTCCATGC ACTCCAACTCCAACAACTCG unc-16 ATTACCTCCACCTGCAAACC CGAATCGACAACCATACTCC unc-17 AGTACGACCGAGTTCTCAGC ACCACTGGAAGACACCACC unc-2 GTCAGATCACTCATGAACTCC ATCCATAGCTCTTGCAACC unc-26 CTATTCCTCTTCGCAAGACC CCCTTCTTTCTCTGATCTTTCC unc-30 CACCATTAACCCACAATCC CAGAGTCGTCTAGCTGACTGC unc-31 AAGCGATCACTGCTCTCC CTCGAATTCTAGCCAATTCC unc-4 ACGATAACCTGTTGATGAACG CACGAATTACATTCCCACC unc-40 CACCAAGTGGAGGACAACC AAACCCAATCCGAGAAGG
unc-42 AATAGCTTCCGCCAAAGG GCAATCGAAGATCCAAATACC unc-5 CATGTACCGTTCCACCTCC CCGTTTTCCACCAGATCC
unc-51 CGAGTCAACGTTTGACCTACC TCCGTCTGCTGATACTTTGC unc-52 CCCACAAACACAGACAATCC ATTCCCCGACATTTCTATCG unc-54 CGGAAAGACTGAGAACACC ACTTCATGTTACCCATGTGC unc-58 TTATGTTTCCCCTACTCGTCC CCATCATCTGCATCGAGC unc-59 GCAGCACAAAGAAAATCC ACACGTCCATGAAGATGC
unc-6 CCATTTCCAATCCCATCC CATCATCCGTCTCGTTTCC
unc-61 ACCGCTTTTCTCATCGTCC TCCTGTTCCCGTTTCTCC unc-64 TCCAGTTAACGATCAGAAAACC AATTCTTTCGCGTGCTCC
unc-7 CAGACATCTCGACAATCTTCC ACTGTGCCAATACAACAATCC unc-71 CTGCAAAAACCCAAAACC CCACACGATTTTGAATAACC unc-75 CCAAAAGACAAAATGGACG ATAAAACTGGGTTTGCTGC unc-76 CAACCTCTCGACTTTGGAACC TCATCTCGATGTGCCAGC
abl-1 CATCATCATCATCTGCACC TAGTTCCCATTCGTCTGG alx-1 TCATGCTGCTGCTAAGAAGG GCTGTTCACTGGACATTCG bath-41 ATTCACAGAGTTTCTATCCACC GCACTCATGTTTATGATTCG bath-42 CATAGGAGAATGCCCTAATCC GATATCGAGATGTTGAAGAAGC bath-43 GACAACAAACAGTTCTCC TGCTCTAAAGACTTCTTCC bath-44 AGAGCAGCTCAAAATACTCG CCAAACCATACATTTCAGC bir-1 GGGACCAAAAAAAAGTCG GGCTGTTAGATTTTGTTGTCG bir-2 GCTCAAAAACTTGAAAGACG GAAATCAATTTCCTGGTGG c04e12.7 GTACAAACTCACACTGTAACG AGCTTTTCCACTTCAACC
cdl-1 TCACCTTCAAAACGTCGTCC TGAGTCCACATCGAACTTTCC ced-3 TCGCTCATTCAGCAAAGC AGAATCCAAGACTGGGAATCC ced-2 GGGAACCACTGGATTAGTGC GAACAAAATGGAATGGACACG
ced-6 ACCAAACATTCCTCCATCC TCCACTTCATTGATCATTGC ced-8 GCTCATCCAAGAAGAACTGG AATGCGACAAGTGTGATAATCC ces-1 CTCTTCTAGTTCGTCAACGTCC ATGTGCGCTCGTAAGTTGC che-13 TGCAAACATAATGGCTGC TATCGAGATAAGGACCAACG cps-6 CAAGCCAGATATCACTTTCC GAGCGAAAATCTCAAGACC crn-2 CGCTTCACAATTTCTCTTCG CGTTCAATGCACATGTAATCG crn-3 CTTTATCATTGATCCCTTCC AAGATGTGTAAGTGCATAGTCC csp-1 CCCACCAAACTAGATGACC CACCCATGTTCAATTTTCC csp-2 ATATCCTGTCTGCCTTGC CTCATCCTTCTTGAATCTCG efl-2 AAGTCAGCAATCACTTGG CATTTGTGAAGAGCAACG fem-1 ACGAAAACATTCAAGGAACACC ATTCATCGCCGACATTGC gla-3 TCTTCGATGCCGTTATCTCC AGACAAAAGTCGATCCATTTCC hsr-9 AACTCACGATACAAACCAAGC ACTCAGCAATTCTAGACTCTGC ikb-1 AAGATGAATCTGGAGAGACTGC CCAATCTTTGCCATTTGC
imb-5 CATATGCTCTCCAAATCACG CATTCCAATGGCAATAATCC math-33 ATGACACCCTGTGCAACC GCATCAAGCTTATCCCATCC mel-26 TATTACCGATATGGCAACTGC TCTTCAACGAGATAAGGAGACC mrt-2 AGGTCTTCAAAACCGTCG ATCGAATTCAAAGTCGAGC unc-1 AAACTCTCGACGACAACG GAATATCCAGCCATAGTCTCC pik-1 CATGAAACGAGAAGTTCTCC CCAATTACATTGCTCACAGC rbx-2 CAATGCAGATTCACAAGAAGG AGAACGGGGTGAAAAATGG
t02c5.1 ATGAGTCCCCTTACGTTACC CGAAACAATTTCGCTTCC T12G3.2 CTATGATCCACCGATTCC CATCCTCTCCACTTGAACC T22H2.5 AACAAGCAATCACAACTCAGC ATCACATTTCCATCTCCATCC T27F7.2 TCATCATCTTCCACTGACACG ATTCCTCCACCACATCTTCC tir-1 TCTCTTCTCTCCTCACTCACC TTTGCCATACATGCATCC Y50C1A.1 AATGAAATTCGAACCCACG GGCTCCATCTTTAGGCACC Y50E8A.9 GACACCAATCGTTCTTCC CATTTGACGAGCAAATCC
ZK1053.5 ATGTGCTCATCGTCAAACG ACAATCAAAAATGTGGCTCC ape-1 TGATCACATTTTCTCCACC TCCATTTCAAGATTCTGTCC
Fwd sequence Rev sequence
msh-2 ACTCGACCGAATCTTCTTCC CAAAATGTCCTGTGCAATAGC nft-1 CCCAACTGCTCTCATTTCC CTCCAGCATTGCTTGAACC par-4 TTTAACGAGCGTCCAACG TCTGAAGCTGAAGCAACACC t13h5.8 TTGTTCAACCCAACCACG CAAACATGATTGGAAGGTATCC t14f9.3 TTGACAAACCACGTTTCC TCCAAATCCTTTCTCCTCC zk686 CTATTCTCTACCATTTCGGACC CCAGCATTCTTTCTATCTTTGC brd-1 GCCACATTTCAACAGAAACC CGACGCTGAAGAATTTCC brc-1 ATCACAGAAACAGTGGCACG CGGGAATACTCGACTTTGG brc-2 TAGTGATCTCAAGAGCAAATCC GATTTCGCACCATTTTCC bub-1 GTTCGCTGAAGTTCTATCTGC TTTCACGAAAACGACACC
daf-18 ACTTCTATCCGAGCCCACG ACCAAACATGACCAATCTTCC cbp-1 CAGTATGCAAGCAACTCATAGC CTCCAGACTGACCCAATCC dic-1 CAACACACTTGGTTTGTGC GATCATATGCGTCGATTCC dog-1 AACGAAACACGTTCTATAGTCC AATTCCGACAGAAATCACC hoe-1 GAACATATGGACAAATGAGAGC TCAACCAACAAATCACATGG
Fwd sequence Rev sequence
maco-1 GAAATCTGAAGCTAGAGTCG GTCAGAATCATTTCCAGC cyp-44A1 CGATCTTCAGTTCAACACG GATCAAATAGCTTGCAAACC E02H1.6 GACGACCGAATATCCTGG GTGCTCATTTTTGAACTCTCC gcs-1 TGTTGTCTTCAAGTCACTTTCC TTTTCTCATCGTCCTGTTCG pqm-1 CAAGAAGTATATCGAACAGACG ACTGCTAAGCAGATTTTTCC
sod-2 GGAGCCTGTAATCAGTCACG TGGCGATCTTCCAAATAGC oxi-1 CCCATCACCAACATCATCC CCGAAATATTGCACATTGC E04F6.5 CGCTGAGTAAATCTGCACG AGCACCCATGACAACACC acdh-2 TGCTCAATCATCAGTCAAACC TTCCCCAAGAACGACTCC F52E1 AGCAACCAGCAAAAAGTCC CGATCCAAGAACAATGAATCC gpd-1 AGTATGACTCGACCCACG GAATGACTTTTCCCACAGC gpd-2 ACATCATCTCCAATGCTTCC TCCGAACTCGTTATCGTACC gpd-4 GATCCGTTTATCACAATCG CATCTCTCCACAGCTTTCC
Table1. Gene primer designed list
The quality and quantity of mRNA is detrimental to this experiment. Wear gloves for all procedures. RNase is everywhere, especially on human skin. Don’t stop until reverse transcription (RT) is done. Change gloves constantly.
2.7 RNA Concentration and Quality
The concentration and purity of total RNA are determined by measuring absorbence at 260 nm (A260) and 280nm (A280) in a spectrophotometer. The quality of total RNA is determined with agarose gel electrophoresis.
2.7.1 RNA extraction protocol
a. Homogenization, use 1 mL Tri-reagent for <0.1 gm tissue, Homogenize until the mixture turns pinky, incubate at RT 5 min, transfer 1mL mixture to 1.5mL tube
b. Phase separation, add 0.2 mL chloroform (pure), vortex, RT 2-15 min
spin, 12K rpm, 4 °C, 8 min, Immediately transfer agueous phase (~ 700 μL) to a new 2.0 mL tube
c. Column purification, Precondition
to agueous phase add equal volume (700 μL) of 70% EtOH Apply 700μL to an RNeasy mini column, spin 1 min at 12K rpm
Repeat Washing
700 μL RW1, 1 min at 12K rpm 500 μL RPE, 1 min at 12K rpm 500 μL RPE, 2 min at 12K rpm
Elution
30 μL RNAse free water, 1 min at 12K rpm Repeat, keep RNA solution on ice
Measure OD260/280 (> 2.0), in phosphate buffer (pH 7.5) (Conversion: 1 OD260 = 30 ug RNA/mL, Calculate yield)
Store unused RNA at –80 °C
Transfer 5 μL in a new tube for RT-PCR and Real-time RT-PCR. Store the rest (45 μL) at 80 °C.
For confirming quality of RNA, we run agarose gel(Fig11) test before experiment of follow-up
2.8 Reverse Transcriptase from RNA to cDNA
cDNA is a more convenient way to work with the coding sequence than mRNA because RNA is very easily degraded by omnipresent RNases. The main reason cDNA is sequenced rather than mRNA. Likewise, investigators conducting DNA microarrays often convert the mRNA into cDNA
2.8.1 Reverse Transcriptase protocol
a. prepare two Annealing mixture: (2X), total RNA (10 ~30 ng)
Oligo dT(12-18) (1μg) 1μL
H2O xμL
Total 31 μL,
65 °C, 10 min, slow cool down to RT (on heat block)
RT reaction, Annealing mix 31μL 5xbuffer 10μL 0.1M DTT 5μL 2 mM dTTP 1μL Rnase inhibitor 1μL SuperScript RT II 1μL Total 50μL
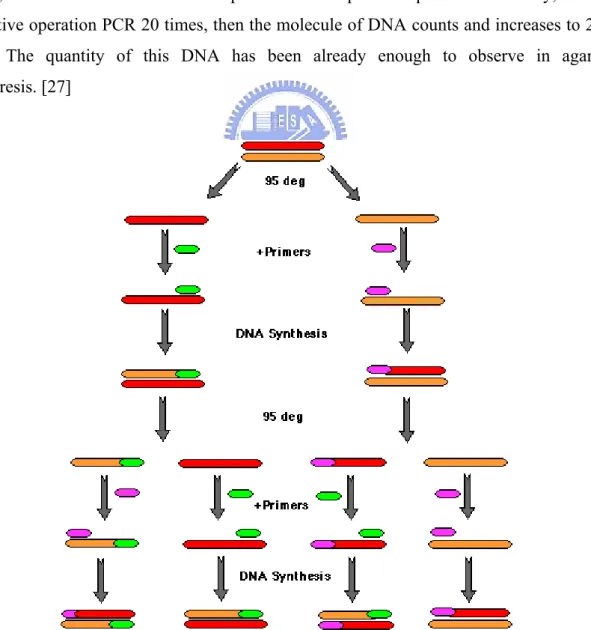
2.9 Polymerase chain reaction (PCR) principle
Polymerase chain reaction (PCR) is a simple and convenient and effective method, it can make DNA increase to above 106X in the micro- test tube. The principle of method is very simple, both ends of bit of DNA increased to want design a leading introduction respectively (forward primer) With putting the introduction instead (reverse primer) .Make it slow and cold with single whiff of goals DNA that has already changed the sex and mate (annealing) And then, utilize DNA to get ferments together (DNA polymerase) Make for the template separately by two shares of goal DNA (template) To formate new DNA share. Picture such via
(1) Denaturation, Make two shares of DNA separated. (2)It is slow and cold to mate and react (annealing), Make the introduction and goal DNA mate. (3)extension),Formate new DNA share. Circulation operate if making by quantity of DNA each time, add every one of, if repetitive operation, many times, calculating with the mathematics formula, the quantity that DNA increases will be 2n, n is the number of times of representative's repetitive operation. In theory, one DNA is like repetitive operation PCR 20 times, then the molecule of DNA counts and increases to 220 = 106 molecule. The quantity of this DNA has been already enough to observe in agarose gel electrophoresis. [27]
2.9.1 PCR protocol
Each gene ddH2O 17.5 l 10X buffer 2.5 l 2mM DNTP 2.5 l cDNA 2 l enzyme 0.5 l primer F 1 l primer R 1 l PCR procedure 95oC 5min 95oC 30s 55oC 30s 72oC 30s repeat 29 times 72oC 10min2.10 Real Time – PCR principle
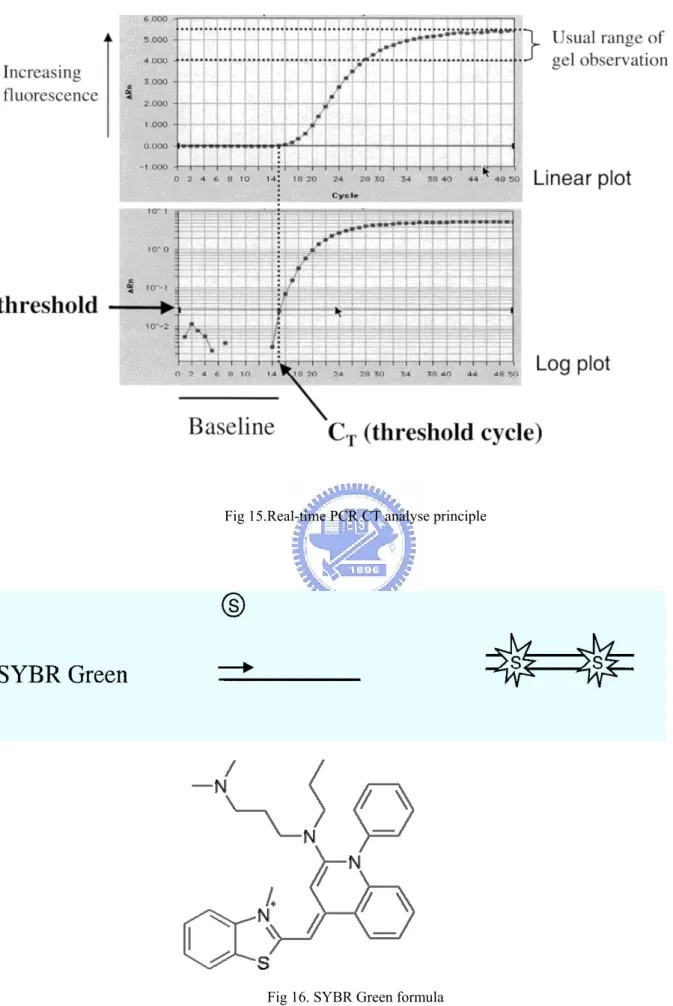
Real-time is an extension of the PCR methodology that has the potential to revolutionize the way in which PCR and quantitative PCR are conducted. In the early 1990s it was shown that the intrinsic 5' nuclease activity of the Taq DNA polymerase could be used to degrade specific fluorigenic probes, thereby allowing for an indirect measurement of the accumulation of the amplicon [24]. Subsequently, it was shown that using a fluorescent dye, the progress of the amplification could be monitored in a closed tube in real time using either a laser
Fig 14. Real-time PCR principle
2.10.1 Relative Quantitation
One simple method for looking at relative differences in expression is the use of the relative CT approach. This approach employs the difference in the CT values obtained for two different sets of samples. Since CT values are obtained during the exponential phase of PCR, it is assumed that at each cycle the number of products are doubling. Assuming that the CT value is reflective of the initial starting copy, a difference of one.
Fluorescence systems used in Real-time-PCR. Systems employ either afluorescent dye such as SYBR Green (S) or employ a fluorescent probe that contains a reporter (R) and a quencher (Q) fluorochrome. Separation of the quencher from proximity to the reporter enables the
Fig 15.Real-time PCR CT analyse principle
2.10.2 Real-time PCR protocol
Each gene ddH2O 10.5 μL TagMix 12.5 μL cDNA 1 μL primer F 1 μL primer R 1 μL Total 26 μL2.11 DNA gel
Agarose gel electrophoresis is an easy way to separate DNA fragments by their sizes and visualize them. It is a common diagnostic procedure used in molecular biological labs.
Electrophoresis:
The technique of electrophoresis is based on the fact that DNA is negatively charged at neutral pH due to its phosphate backbone. For this reason, when an electrical potential is placed on the DNA it will move toward the positive pole:
2.11.1 DNA gel protocol
1. Add the appropriate volume of 1xTAE to a sterile container 2. Add the appropriate amount of agarose
3. Heat in a microwave with the lid slightly loosened until boiling (about 2 minutes on full power) 4. Allow to cool to 'hand warm' (50-60。
C) e.g. by running under a cold tap 5. Add the appropriate amount of ethidium bromide .
6. Pour the gel into the gel casting tray 7. Remove any bubbles in the gel
8. Allow the gel at least 40 minutes to set (or less if put in a fridge) ensuring that the gel casting tray is level and undisturbed
9. When the gel has set remove the combs and casting gates and transfer to the gel tank 10. Mix 5 volumes of PCR product with 1 volume of 6x gel loading buffer
11. Load samples into the wells in the gel
12. Load 1kb ladder into at least one well in each row
13. Run the gel at the appropriate voltage for about 30 minutes 14. Photograph the gel under ultraviolet light
Chapr 3
Result & Discussion
3.1 C.elegans behavior
Use a kind of new quantitative way in the experiment that C.elegans behavior observes
Op 50 Ecoli is scribbled on agar plate, because it shed the orbits of sports when C.elegans creeps. We use very small graph paper cushion under agar plate.We time 1 min to calculate the total length of orbit that C.elegans creep at this moment. Because we find a simple fact in the past research of our laboratory. The magnetic field could reduce the C.elegans crawling the speed.
The following charts explain observation of C.elegans' behavior
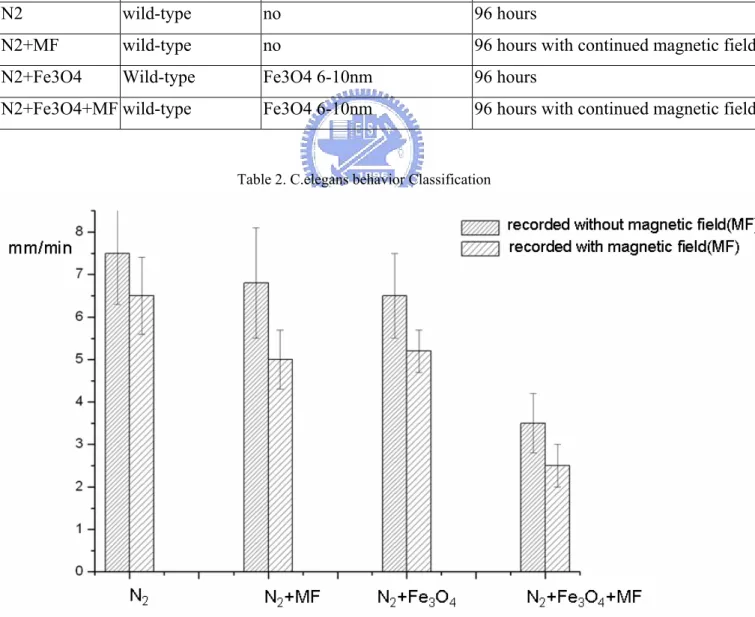
We use N2 C.elegans. We observed the C.elegansto creep and slackening. Divided into four groups as follows
C.elegans strain additive nanoparticles culture environment
N2 wild-type no 96 hours
N2+MF wild-type no 96 hours with continued magnetic field N2+Fe3O4 Wild-type Fe3O4 6-10nm 96 hours
N2+Fe3O4+MF wild-type Fe3O4 6-10nm 96 hours with continued magnetic field
Table 2. C.elegans behavior Classification
The above is taken a sample at random by 25, ρ<0.05 .We find the magnetic field has lasting influence on C.elegans. The magnetic field makes the behavior slacken. Add magnetic nanoparticles increase the disparity of influencing. Assigning to four groups is for proving the prescroption and nanoparticle of the magnetic field act on together. There is an influence on influence on the magnetic field in the C.elegans for a long time
For example 2 groups 1.N2 and N2+MF
1.N2 slows down, after culturing 96 hours with continued magnetic field.
If we recorded with magnetic field, we have a fack that N2 slow down acutely . There is transient influence on influence on the magnetic field in the C.elegans during our experiment. when we take magnetic field off, the transient influence disappear. However, the long influences on slowing down still exist. The result is exciting. We add super-paramagnetic nanoparticles in our experiment. The result changes greater
3.1.1 Video recorded
We use the record system made by oneself to analyse the behavior of the C.elegans.The original text one is a video. In order to appear convenient, we use and pursue to explain in succession.
The order is 1 → 2 → 3 → 4 →5
↓
6 → 7 → 8 →9 →10
C.elegans behavior speed
a.Control-N2>b.N2+Fe3O4>c.N2+magnetic field>
d.N2+Fe3O4+magnetic field 48hr>e.N2+Fe3O4+magnetic field 96hr
b.N2+Fe3O4
c.N2+magnetic field
d.N2+Fe3O4+magnetic field 48hr
e.N2+Fe3O4+magnetic field 96hr
3.2 Super-paramagnetic nanoparticles in C.elegans
3.2.1 Super-paramagnetic nanoparticles-Fe3O4
Show in documents before this it is nonpoisonous.We buy super-paramagnetic nanoparticles from Taiwan Advanced Nanotech Inc Amino-TANBeads / U118 Magnetic beads-Amino terminated (-NH3)
Carboxyl-TANBeads /U128 Product list Magnetic beads-Carboxyl terminated (-COOH)
3.2.2 Optics microscope picture
In order to confirm that really there is FE3O4 that we add in nematode's body
We use the optics microscope to observe the state after 3hr feeding. The following of the result
Control
Fig 24. N2 optics microscope picture
The background light source is unavoidable.
C.elegans under the optics microscope is clear and is very easy to observe: We could find nanoparticle appear in the throat or alimentary canal.
Experiment
3.3 C.elegans SDS-page
Fig 26. C.elegans SDS-page
Function paper of the C.elegans and magnetic field .Exposure of C.elegans to extremely low frequency high magnetic fields induces stress responses. A number of studies suggest that the expression of heat shock protein induced by heat stress is associated with protection. So we follow it.
However, the result is not good. BSA is mark and molecular weight is 69kb. It is lower than 69kb that we can infer the molecule in the red frame bright. 3 groups on the left are all N2+Fe3O4. On the right is normal N2. We can guess at least that adding Fe3O4 will not influence proteins. (Whether data behind is it join Fe3O4 can take to gene some influence)
3.4 PCR data
3.4.1 Gene number
For the convenience on the experiment, we number the gene.
Follow-up experiment that it is here that Arabic numeral represents the gene.
1 unc-1 20 unc-52 39 c04e12.7 58 math-33 77 t14f9.3 96 oxi-1 2 unc-103 21 unc-54 40 cdl-1 59 mel-26 78 zk686 97 E04F6.5 3 unc-112 22 unc-58 41 ced-3 60 mrt-2 79 brd-1 98 acdh-2 4 unc-108 23 unc-59 42 ced-2 61 K03E6.5 80 brc-1 99 F52E1
5 unc-115 24 unc-6 43 ced-6 62 pik-1 81 brc-2 100 gpd-1 6 unc-119 25 unc-61 44 ced-8 63 rbx-2 82 bub-1 101 gpd-2 7 unc-129 26 unc-64 45 ces-1 64 t02c5.1 83 daf-18 102 gpd-4 8 unc-13 27 unc-7 46 che-13 65 T12G3.2 84 cbp-1 103 hsp16 9 unc-16 28 unc-71 47 cps-6 66 T22H2.5 85 dic-1 104 hsp70 10 unc-17 29 unc-75 48 crn-2 67 T27F7.2 86 dog-1 105 hsp90 11 unc-2 30 unc-76 49 crn-3 68 tir-1 87 hoe-1 106 act-1 12 unc-26 31 abl-1 50 csp-1 69 Y50C1A.1 88 maco-1 107 age-1 13 unc-30 32 alx-1 51 csp-2 70 Y50E8A.9 89 cyp-44A1 108 dif-2 14 unc-31 33 bath-41 52 efl-2 71 ZK1053.5 90 E02H1.6 109 dif-16 15 unc-4 34 bath-42 53 fem-1 72 ape-1 91 gcs-1 110 L18 16 unc-40 35 bath-43 54 gla-3 73 msh-2 92 pqm-1 111 L21-1 17 unc-42 36 bath-44 55 hsr-9 74 nft-1 93 skn-1 112 L21-2 18 unc-5 37 bir-1 56 ikb-1 75 par-4 94 smk-1
19 unc-51 38 bir-2 57 imb-5 76 t13h5.8 95 sod-2
Table.3 Gene number list
3.7.1 .PCR electrophoresis
The following is a result of PCR. It proves the magnetic field will cause the change on the gene. Base on observation in C.elegans behavior in the front, we know Fe3O4 + the magnetic field synergy a largest influence on C.elegans. Group Control is normal N2. Group Experiment feeds N2 which eats Fe3O4 + magnetic field function. So rough classification is essential .It is not quantitative that PCR behaves. We can fast to find which gene have influence with law this. On the right the
Fig 27.Gene PCR gel 1-25
From 1 to 25, the genes about behavior behave. Seeming to be data has not presented the difference on the result. Of possible .It is a lot of factors will be influenced between primer design and temperature.
Fig 28.Gene PCR gel 26-50
From 26 to 50, later 30 apoptosis was correlated with. Later it was the place where we are relatively interested in. The experiment is brighter than the control and more in most places. This we do not confirmed. The expression of PCR is the showing uncomfortable ration.
Fig 29.Gene PCR gel 51-74
From 50 to 74 .almost are apoptosis related gene and cancer related gene. In contrast, impaired apoptosis may be a significant factor in the etiology of such diseases as cancer, autoimmune disorders, and viral infections.
Fig 30.Gene PCR gel 75-99
From 75 to 99, he left one side of something is that cancer is correlated with. The right one side of something is that oxidation is correlated with. From 1 to99, we find that deduct background he difference of every gene is diminished or has no way to assay. Some are not even believable. So except traditional PCR, we need good tools to prove the difference that the gene expression.
Fig 31.Gene PCR gel 100-109
From 100 to 109, C.elegans and magnetic field function in possible relevant gene studies.
In the research before this, the magnetic field think that correlate with HSPs and C.elegans is studied most frequently usedly in life span .So this group hopes to reappear over forefathers' research.
Fig 32. Repeat gene PCR gel 76-96, L18, L21
We can affirm even more with this picture the magnetic field must influence with the gene relevantly. L18, L21 on the right is control gene. If the result it is expected, the luminance of group the experiment and group the control are the same. We can confirm 2 genes expression are the same But other groups show different results. Seem to have an effect in magnetic field.
3.5 Real-time PCR data
Real-time PCR is very sensitive. The repetition which wants 3 times is believable. The first data is review, in order to test the stability of data. Because real-time PCR cost is expensive, effective DATA appears as follows. We test all genes first.
3.5.1 Real-time PCR system
The new MyiQ real-time PCR detection system offers an affordable alternative for the detection of common green fluorescent dyes such as FAM and SYBR Green I. This system interfaces directly with the iCycler thermal cycler, offering superior features such as thermal gradient and Peltier-effect driven performance. The MyiQ real-time PCR detection system is a perfect solution for those just getting started with this technology as well as those looking for additional instruments to handle increasing routine assay demands. This system was developed by the same experts at Bio-Rad who have pioneered performance in real-time PCR. The MyiQ delivers the same excellent-quality data as the iCycler iQ real-time detection system.
3.5.1.1 Real –time PCR procedure
3.5.2 Gene expression
We use real-time PCR to prove gene expression. The following axes of ordinates of picture (Y axle) Show the disparity that the gene expression. Disparity that the gene expression is a sub number of 2
Gene expression = 2
nNow we give an example in 3.5.3 Fist real-time PCR test(flow)
For example1, gene number 31 Y axle is similar -3
Gene expression = 2
-3= 8
-1We can say in this gene number 31 gene expression the experiment is 8 times lower than the control. So gene number 31 Gene expression was reduced in the mechanism of testing at this moment. And it has reduced by 8 times
For example2, gene number 75 Y axle is similar 14
Gene expression = 2
14= 16384
We can say in this gene number 75 gene expresssion in the experiment is 16384 times higher than thecontrol. So gene number 75 Gene expression increased in the mechanism of testing at this moment. And it has increased 16384 times.
3.5.3 Fist time real-time PCR test
control N2
experiment N2+Fe3O4+MF
The axes of ordinates of chart(Y axle) are t △gene expression(the experiment-the control) The axes of horizontal of chart(X axle) are the gene name serial numbers
The result is not believable, because it is only work one time.
The positive number expresses the experiment group > the control. The negative number expresses the experiment group < the control
3.5.4 Real-time PCR data believable
The result is believable. It is three time replay.Real-time PCR is very sensitive, so we must utilize knowledge, experience and effective tool to get rid of useless data. A simple method is to see whether PCR Amp/Cycle and Melt Curve accord with expectancy.
Prove by following 2 Graph. We adopt just with PCR curve intact and sharp Melt Curve. Because data is numerous, we got rid of no influencing and data disorderly. The following data is all covering triply, the credibility is very high
PCR Amp/Cycle Graph for SYBR-490
Fig 35. PCR Amp/Cycle Graph for SYBR-490 Melt Curve Graph for SYBR-490
3.5.4.1 Ced family gene expression
Fig 37.Ced real-time PCR data to gene expression
Ceds are very famous for apoptosis research gene . The control is normal N2
The experiment 1 is N2 +Fe3O4
The experiment 2 is N2 +Fe3O4+mabnetic field (MF)
We can observe that all gene expression increase with magnetic field obviously. Though add Fe3O4 to gene expression has influence, but we can confirm the magnetic field has really influenced gene expression even more.
3.5.4.2 Other apoptosis related gene expression
Fig 38.Other apoptosis related gene real-time PCR data to gene expression
The control is normal N2 The experiment 1 is N2 +Fe3O4
The experiment 2 is N2 +Fe3O4+mabnetic field (MF) These apoptosis related genes are all very famous. Take abl-1 as an example:
A very obvious one that adds Fe3O4 to C.elegans is not affected. But gene expression is very obvious under magnetic field function. Magnetic field function disparity is reached 25 = 32 times.
3.5.4. Some cancer related gene expression
Fig 39.Some cancer related gene real-time PCR data to gene expression
The control is normal N2 The experiment 1 is N2 +Fe3O4
The experiment 2 is N2 +Fe3O4+mabnetic field (MF
We are very sorry about that it is not easy to look over t the chart small very much. But we can still perceive the magnetic field has really influenced gene expression.
3.6 C.elegans GFP marked
Fig 40. GFP mark signal
From CGC mutant C.elegans, some strain has GFP mark; .liko cbp-1.Cbp-1 has GFP mark in pharyngeal. We culture cbp-1 C.elegans with the magnetic field 96 hr (right) and with no magnetic environment (left).
Fig 41.GFP signal decrease with magnetic field
We find an interesting thing that pharyngeal GFP signal will be reduced acting on continuously with magnetic field. Though CBP-1 can change with the magnetic field, pharyngeal GFP signal is not reporter gene GFP signal. Although the cbp-1 gene expression decrease with magnetic field in our experiment, it can't be proved while making people interesting like this.
3.7 C.elegans mutant behavior & real-time PCR data
Fe3O4 ultra paramagnetic nanoparticles influence gene expression and strengthen the impact on gene expression of the magnetic field. But this ultra paramagnetic nanoparticles has spontaneous magnetic field. In order to distinguish the spontaneous magnetic field and the magnetism field influence. We join 2 kinds of nonpoisonous nanoparticles-Fe2O3, TiO2...Fe2O3andTiO2.diameter are (40nm) slightly biger than Fe3O4 (10nm).
Fe2O3 is compared with Fe4O3; they are all oxides of the iron. The difference is the ultra paramagnetic in Fe3O4 has spontaneous magnetic fields. The same one they are attracted by magnetic field. : They all move with the direction of the magnetic field. Join Fe2O3 in order to prove what is influenced is in the experiment. It was not only a spontaneous magnetic field that was influenced, the more important thing environment magnetic field.
TiO2 is compared with Fe4O3. They are all nonpoisonous nanoparticles. Fe3O4 ultra paramagnetic nanoparticles influence gene expression and strengthen the impact on gene expression of the magnetic field. Nanoparticle influence gene expression or environment magnetic field. This is important thing. TiO2 is useful for riding of the influence of the iron oxide and distinguishing nanoparticles influences.
We request strains from CGC
C.elegans mutant gene CGC strain C.elegans mutant gene CGC strain ced-3 MT1522 C04E12.7 BC13266
ced-6 MT4970 ces-1 DC1079
abl-1 XR1 math AA278
che-13 CB3323 brc-2 DW104
inft-1 VC909 cyp-44a1 VC206
par-4 KK184 gcs-1 VC337
cbp-1 VC1006 F52E1 BC10281
unc-17 LX929 unc-108 VC829
unc-30 EW45 unc-119 XA3504
Now we use 3 new mutant strains, ced-3 MT1522, ced-6 MT4970, cbp-1 VC1006 to prove our result.Ced-3 and ced-6 are apoptosis related gene. Cbp-1 is cancer related gene. They are all deficient mutant.Ced-3 and ced-6 is in the same pathway in apoptosis. We find apoptosis deficient mutant is insensitive to magnetic field.
Mutant location genomic environs
Ced-3 MT1522
Ced-6 MT4970
Cbp-1 VC1006
3.7.1 Ced-3 MT1522
3.7.1.1 Ced-3 mutant behavior
Fig 43. Ced-3 mutant behavior with magnetic field
The above is taken a sample at random by 25, ρ<0.05.Recording mode is as same as before utilizing. We compare normal N2 with ced-3 mutant C.elegans. The result is that ced-3 mutant C.elegans is not sensitive to the magnetic field.
3.7.1.2 Ced-3 mutant real-time PCR data
Fig 44. Ced-3 mutant real-time PCR data to gene expression
The type adds the kind of nanoparticles above the chart (bar1 has no nanoparticle)
It is three time replay. Different display ways before following. : Question that we melt simply. Whether only the magnetic field will have changes to ced-3 mutant C.elegans. The result is excited, not adding any nanoparticle, (bar1)
Gene expression with magnetic field increase more than 5 times of Gene expression without magnetic field. This result expresses magnetic field and ced-3 positive correlation.Ced-3 is important apoptosis pathway. We can suppose boldly the magnetic field correlate with apoptosis.
The magnetic field causes apoptosis.
The magnetic fields still change gene expression and it is positive correlated. Though seem to add nanoparticles (Fe3O4 Fe2O3 TiO2) reduce the difference. We do not care about this. Because on generally speaking, it contain inside great and complicated problem that are not solved.
3.7.2 Ced-6 MT4970
3.7.2.1 Ced-6 mutant behavior
Fig 45. Ced-6 mutant behavior with magnetic field
The above is taken a sample at random by 25, ρ<0.05 Recording mode is as same as before utilizing.
we compare normal N2 with ced-6 mutant C.elegans.
Two of their results are similar. The possible reason is as follows 1. Ced-6 mutant damage is not serious
2. Gene relevant degree is not enough 3. Gene pathway not related directly
Behavior datas show ced-6 mutant and magnetic field function have no relation between with themselves. The result accords with the expectancy of apoptosis pathway
3.7.2.2 Ced-6 mutant real-time data
Fig 46. Ced-6 mutant real-time PCR data to gene expression
If C.elegans gene related to phagocytosis mutant in the body, liko ced-6, may escape death. Except that the passive role with death cell's skeleton of a corpse of phagocyting and is not merely acted and moved around the cell. Even cause the initiative assailant of apoptosis [31].
Ced-3 and ced-6 are not in same pathway , although they are apoptosis related. The result shows the lower gene expression with magnetic field function.
3.7.3 Cbp-1 VC1006
3.7.2.1 Cbp-1 mutant behavior
Fig 48. Cbp-1 mutant behavior with magnetic field
The above is taken a sample at random by 25, ρ<0.05. Cbp-1 mutant is compared with N2.
We look cbp-1 mutant C.elegans is more sensitive to the magnetic field instinctively. This kind of method instinctively lacks the scientific idea.
N2 speed under magnetic field function reduces almost 3 mm/min. (N2 vs. N2+MF from 7 to 4) Cbp-1 mutant speed under magnetic field function reduces almost 3 mm/min. (cbp-1 vs. cbp-1+MF from 5.5 to 2.5).
Their disparity is all 3. We can not put the final conclusion that cbp-1 mutant C.elegans is more sensitive to the magnetic field instinctively.
3.7.2.2 Cbp-1 mutant real-time data
Fig 49. Cbp-1 mutant real-time PCR data to gene expression
Over 80% of the cancers known produce, because p53 is lost badly at present. P53 is important, because it has 3 functions:
1. Order cells not to grow again, is engaged in the work of mending damaged DNA attentively (growth arrest).
2. When being unable to mend, order cells to commit suicide.
3. Suppress the blood vessel hyperplasia (anti-angiogensis), Supply the cancer cell with a nutrient, and make the cancer cell unable to grow up, shift.
Produce the cancer, but destroy p53 .
Celegans cbp-1 is similar to human P300/CBPP300/CBP is activating of p53, by combining with p53 N end, and 382 acetylating lysine, increase p53 activate the ability that the gene expression. [32] Cbp-1 gene expression accords with expectancy. Cbp-1 gene expression is cancer negative related.The result infers the magnetic field correlate with cancer genes.