R E S E A R C H A R T I C L E Open Access
The genome and occlusion bodies of marine
Penaeus monodon nudivirus (PmNV, also known as
MBV and PemoNPV) suggest that it should be
assigned to a new nudivirus genus that is distinct
from the terrestrial nudiviruses
Yi-Ting Yang1,2, Der-Yen Lee3, Yongjie Wang4,5, Jer-Ming Hu6, Wen-Hsiung Li7,8, Jiann-Horng Leu9,10,
Geen-Dong Chang11, Huei-Mien Ke7,12, Shin-Ting Kang1,2, Shih-Shun Lin13, Guang-Hsiung Kou2*and Chu-Fang Lo1,2,14*
Abstract
Background: Penaeus monodon nudivirus (PmNV) is the causative agent of spherical baculovirosis in shrimp (Penaeus monodon). This disease causes significant mortalities at the larval stage and early postlarval (PL) stage and may suppress growth and reduce survival and production in aquaculture. The nomenclature and classification status of PmNV has been changed several times due to morphological observation and phylogenetic analysis of its partial genome sequence. In this study, we therefore completed the genome sequence and constructed phylogenetic trees to clarify PmNV’s taxonomic position. To better understand the characteristics of the occlusion bodies formed by this marine occluded virus, we also compared the chemical properties of the polyhedrin produced by PmNV and the baculovirus AcMNPV (Autographa californica nucleopolyhedrovirus).
Results: We used next generation sequencing and traditional PCR methods to obtain the complete PmNV genome sequence of 119,638 bp encoding 115 putative ORFs. Phylogenetic tree analysis showed that several PmNV genes and sequences clustered with the non-occluded nudiviruses and not with the baculoviruses. We also investigated the characteristics of PmNV polyhedrin, which is a functionally important protein and the major component of the viral OBs (occlusion bodies). We found that both recombinant PmNV polyhedrin and wild-type PmNV OBs were sensitive to acid conditions, but unlike the baculoviral OBs, they were not susceptible to alkali treatment.
Conclusions: From the viral genome features and phylogenetic analysis we conclude that PmNV is not a baculovirus, and that it should be assigned to the proposed Nudiviridae family with the other nudiviruses, but into a distinct new genus (Gammanudivirus).
Keywords: PmNV, Genome, Baculovirus, Nudivirus, OBs, Polyhedrin
* Correspondence:ghkou@ntu.edu.tw;gracelow@mail.ncku.edu.tw
2Department of Life Science, National Taiwan University, Taipei, Taiwan
1Institute of Bioinformatics and Biosignal Transduction, College of Bioscience and Biotechnology, National Cheng Kung University, Tainan, Taiwan Full list of author information is available at the end of the article
© 2014 Yang et al.; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly credited. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
Yang et al. BMC Genomics 2014, 15:628
http://www.biomedcentral.com/1471-2164/15/628
Background
Spherical baculovirosis is a shrimp disease that was first observed in Taiwan, and it was also the first reported viral disease from Penaeus monodon [1]. The viral patho- gen that causes this disease is now widely distributed along the Indo-Pacific coasts of Asia, and it infects a range of penaeid shrimps. The virus is a rod-shaped, singly enveloped and occluded large circular dsDNA virus that replicates within the nucleus, and it targets several organs, including the hepatopancreatic tubule epithelium and duct epithelium of postlarvae, juveniles and adults, and the anterior midgut epithelium of very young postlarvae [2].
When it was first discovered in 1981, it was thought to be a baculovirus because of the structure of its occlusion bodies as revealed by electron microscopy [1]. Two years later, it was designated monodon baculovirus (MBV) [2], and this name is still commonly used today. One decade after its discovery, Mari et al. showed that each envelope contained a single nucleocapsid, and that numerous virions were included within each occlusion body. Mari et al. therefore proposed that MBV be assigned to the subgenus SNPV with the name PmSNPV [3]. Later still, from 2005 to 2011, in the 8threport of the International Committee on Taxonomy of Viruses (ICTV), Penaeus monodon nucleopolyhedrovirus (PemoNPV) was listed as a“tentative species” in the Nucleopolyhedrovirus genus.
Meanwhile, even though the virus forms occlusion bodies, in 2009, Wang and Jehle used a molecular phylogenetic analysis of six viral genes and supermatrix methods to propose that this virus should to be re-assigned to the genus Nudivirus. They also proposed that it be renamed to P. monodon nudivirus, PmNV [4]. Although the taxo- nomic status of this virus is still in dispute, and it is not included in the 9th ICTV report (2012), the evidence we present here supports this proposed reassignment, and we will use this new name throughout the present manuscript.
Before the present study, only a 22.8 Kb partial PmNV genome sequence was available in GenBank. To further clarify its phylogenetic status, we used an NGS (Next Gen- eration Sequencing) platform to determine the complete genome sequence of PmNV, and then analyzed and com- pared its genomic features with other closely related species. We also investigated the unique properties of the polyhedrin that forms PmNV’s occlusion bodies.
Our results suggest that PmNV is not a baculovirus, and we propose that it should be assigned to a third genus, Gammanudivirus, within the newly proposed Nudiviridaefamily [5].
Results and discussion Sequencing of the PmNV genome
The complete PmNV genome was sequenced by an Illumina Miseq sequencer using the paired-end method.
High-throughput sequencing was performed twice to compare between different sample preparation methods and sequencing conditions. The longest contigs from the first and the second sequencing were 119,426 and 119,128 nt, respectively. However, high-throughput NGS sequencing can produce nucleotide errors, while the short sequences used by NGS can lead to repetitive errors in assembly. Therefore, based on the longest contig, we designed 374 primers (Additional file 1: Table S1) and used Sanger sequencing to recheck sequences that had unreliable signals. A total of 754 runs were assembled into a single contiguous sequence with a size of 119,638 bp. Comparisons of this sequence with the two high-throughput contigs found a 1 nt insertion and a total of 211 nt deletions in the contig from the first high-throughput sequencing, and 3 nucleotide errors and 556 nt deletions in the contig from the second high- throughput sequencing (Additional file 2: Table S2).
Although the second sequencing produced about 3 × more data than the first, its mapping rate was about 3 × lower, and overall, the mappable reads were almost the same (Additional file 3: Table S3). Since these two high-throughput sequencings were performed on samples of viral genomic DNA with different purity and concen- tration, the similarity of the two sequencing results there- fore suggests that good results can be obtained even when the sample is contaminated with host DNA as long as there is sufficient coverage for accurate assembly.
We conclude that the complete circular genome of PmNV is 119,638 bp in size. This length is in good agree- ment with the 80-160 kb estimated by BamHI digestion [3]. The PmNV genome has a G + C content of 34.5%, which is lower than the G + C content of the type species of the baculoviruses (AcMNPV) and all but one of the four nudiviruses (Table 1).
Repetitive sequences in the PmNV genome
Homology regions (hrs) are an important feature in the genomes of many dsDNA viruses. These regions, which are AT-rich and consist of direct repeats with imperfect palindrome sequences, play a central role in replication [11-13] and also act as transcriptional enhancers [14].
hrs vary in length, sequence and copy number between species, and most of the baculoviruses have several hrs distributed around their respective genomes. Notable exceptions include Trichoplusia ni single nucleopolyhe- drovirus (TnSNPV) [15], Chrysodeixis chalcites nucleo- polyhedrovirus (ChchNPV) [16] and Agrotis segetum granuloviruses (AgseGV) [17], none of which have any hrs. To date, no hrs have been found in any of the fully sequenced nudiviruses, ie. HzNV-1 (Heliothis zea nudi- virus 1) [6], HzNV-2 (Helicoverpa zea nudivirus 2) [7], GbNV (Gryllus bimaculatus nudivirus) [8] and OrNV (Oryctes rhinoceros nudivirus) [18]. The PmNV genome
Yang et al. BMC Genomics 2014, 15:628 Page 2 of 24
http://www.biomedcentral.com/1471-2164/15/628
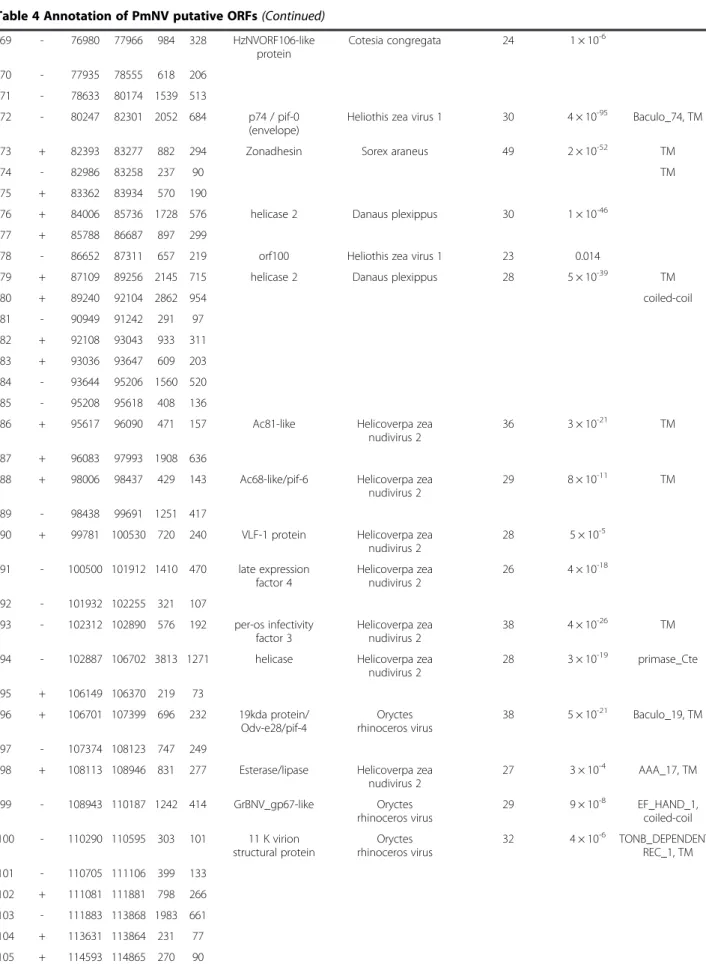
also does not include any hrs. Like the nudiviruses, however, it does include a number of tandem direct repeats (dr; Additional file 4: Table S5). Four of these repeats (dr2-5) cluster together between locations 33,271 and 33,962 in the PmNV genome, which is predicted to be a non-coding region (Figure 1). The repeat unit, copy number and total length of the repeats range from 3 to 42 bp, 2.0 to 8.7 and 26 to 284 bp, respectively, which means that repeat sequences are less abundant in PmNV than in, for example, HzNV-2. Taken together, these results suggest that PmNV and the nudiviruses
might have a replication origin pattern which is distinct from the baculoviruses.
ORF prediction
The ORFs predicted by DNAMAN and the online pro- grams GeneMarkS, GLIMMER3 and FGENESV0 are shown in Additional file 5: Table S4. The A of the ATG start codon of the PmNV001 (polyhedrin gene [19]) was defined as nucleotide 1 of the PmNV genome. To reduce the number of false positives and improve accuracy, only those ORFs that were predicted by at least two programs Table 1 Comparisons of the genome of PmNV and various dsDNA virus
Virus Genome size (bp) No. of ORF G + C content (%) Gene density (kb) Reference
PmNV 119,638 115 34.5 1.04 This study
HzNV-1 228,108 155 41.8 1.47 Cheng et al. [6]
HzNV-2 231,621 113 42.0 2.04 Burand et al. [7]
GbNV 96,944 98 28.0 0.99 Wang et al. [8]
OrNV 127,615 139 42.0 0.92 Wang et al., [9]
AcMNPV 133,894 156 40.0 0.73 Ayres et al., [10]
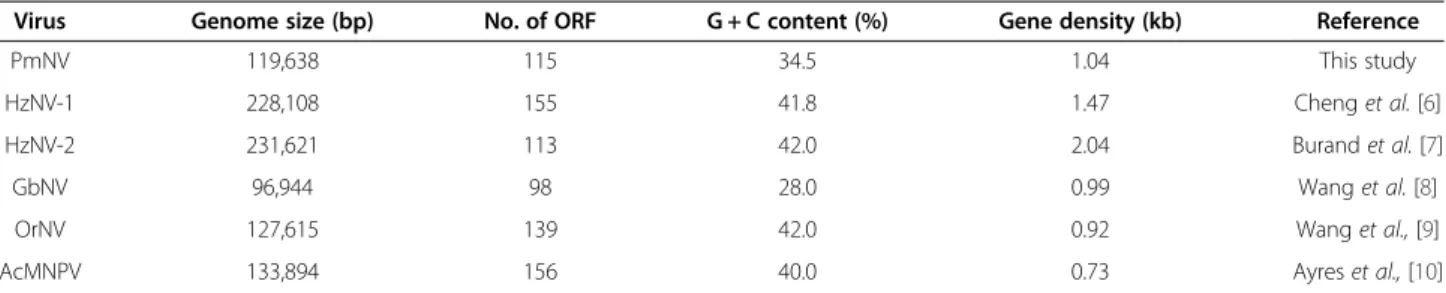
Figure 1 Circular map of the PmNV genome. Purple represents the 60 forward strand ORFs and blue represents the 55 reverse strand ORFs.
Red represents the 10 direct repeat (dr) regions, which are dispersed around the genome. The innermost circle shows GC skew, which indicates possible locations of the DNA leading strand, lagging strand, replication origin, and replication terminal during DNA replication. Below average GC skew is light orange and above average dark orange. The next innermost circle is a GC plot, with light green representing below average GC content, and dark green indicating above average GC content.
Yang et al. BMC Genomics 2014, 15:628 Page 3 of 24
http://www.biomedcentral.com/1471-2164/15/628
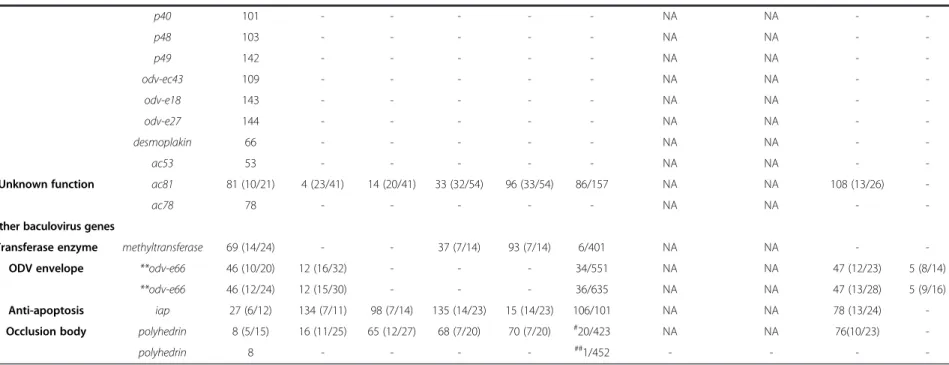
or which had an E-value of less than 1 in the BLASTP search were selected for further analysis. In total, 60 pre- dicted ORFs were found on the forward strand and 55 predicted ORFs on the reverse strand, with sizes ranging from 144 bp to 7.5 kb (Figure 1). The average length of the predicted ORFs was about 990 bp and the gene density was 1.04 per kb. Eight of the 115 putative ORFs (PmNV003, PmNV006, PmNV016, PmNV033, PmNV049, PmNV067, PmNV073 and PmNV113) are similar to proteins found in eukaryotic organisms, while 45 ORFs (39% of 115 putative ORFs) have significant homologies to genes found in other dsDNA viruses (Table 2). Homolo- gous genes were found especially in two closely related nudiviruses with 93% sequence identity, HzNV-1 and HzNV-2 [7]. By comparison, the Baculoviridae share 37 core genes among the Alphabaculovirus, Betabaculo- virus, Gammabaculovirusand Deltabaculovirus [20], 20 of which are also homologous to nudivirus genes [18].
Except for vp39, PmNV has the same homologous genes as the nudiviruses (Table 2). In addition to the nudiviruses and baculoviruses, PmNV also shares some homologous genes with the bracoviruses and hytrosa- viruses, two viral groups that are closely related to the nudiviruses and baculoviruses (Table 3).
Gene parity analysis
Gene parity plots that compared the gene organization of PmNV’s predicted ORFs with those of two representative nudivirus species, HzNV-1 and OrNV, showed no signifi- cantly similar pattern of gene location within the respect- ive genomes (Additional file 6: FigureS1). However, gene parity is not generally conserved in either baculoviruses or nudiviruses, and we note that the conserved gene cluster of helicase and pif-4 (19 kDa) that is found in the known nudiviruses [18] was in fact also detected in the PmNV genome (PmNV094 and PmNV096) though in opposite orientation (Additional file 7: FigureS2). This suggests that PmNV has a closer evolutionary relationship to the nudi- viruses than to other large dsDNA viruses.
Functional and phylogenetic analysis of PmNV putative ORFs that are homologous to baculovirus (AcMNPV) genes Enzymes involved in DNA replication
The PmNV genome includes two ORFs, PmNV005 (dna polymerase) and PmNV094 (helicase), that are involved in DNA replication and play essential roles in DNA polymerization and DNA unwinding, respectively. Two other enzymes LEF-1 and LEF-2, which are factors essen- tial for primase activity in baculoviruses, did not have any homologs in PmNV.
PmNV005 was predicted to belong to the DNA poly- merase type-B family, and it has the POLBc signature from 151-304 aa (Pfam: E = 1.5 × 10-19). The closest matches for PmNV005 are the DNA polymerases of
HzNV-1 and HzNV-2, which both have 26% amino acid identity (BLASTP: E = 2 × 10-102; Table 4). DNA polymer- ase is a key enzyme in virus taxonomy, and it has often been used to construct phylogenetic trees of DNA viruses.
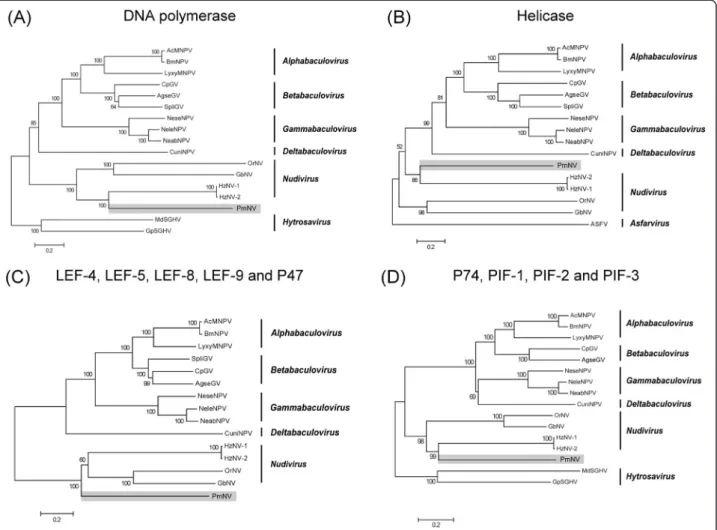
In Figure 2A, the putative PmNV DNA polymerase protein sequence (PmNV005) was aligned with DNA polymerase sequences from closely related dsDNA viruses, including baculoviruses, nudiviruses and hytrosaviruses (as an outgroup), and subjected to phylogenetic analysis.
Each node had a high bootstrap support percentage, indi- cating that this tree was reliable. PmNV was on a branch that was distant from the baculoviruses and closely related to the nudiviruses, which is consistent with the result reported by Wang and Jehle [4] for the partial PmNV gen- ome sequence. Except for HzNV-1 and HzNV-2, the tree lengths show that there are greater evolutionary distances between the nudiviruses than between the baculoviruses.
PmNV094 showed 28% identity with helicase from HzNV-1 and HzNV-2 (BLASTP: E = 3 × 10-19; Table 4) and was predicted to contain the primase_Cte domain (CDD: E = 4.99 × 10-7). In the phylogenetic tree based on the helicase protein sequences of baculoviruses and nudi- viruses, which also had a high bootstrap support percentage at each node, PmNV again formed a cluster with HzNV-1 and HzNV-2 (Figure 2B). As with dnapol, evolutionary distances between the nudiviruses and PmNV are still greater than those within the baculoviruses.
RNA polymerase subunits and an RNA transcription initiation factor
Five putative genes, PmNV014 (p47), PmNV023 (lef-8), PmNV052 (lef-5), PmNV058 (lef-9) and PmNV091 (lef-4), are homologous to those involved in baculovirus tran- scription. These genes are expressed in the late or very late infection stage, and they play different transcriptional roles. LEF-5 is a transcription initiation factor [21], while P47, LEF-4, LEF-8 and LEF-9 are RNA polymerase sub- units. Although p47 and lef-9 are fused into a single gene in HzNV-1 and HzNV-2, all five of the genes in the transcrip- tional group are conserved between the baculoviruses and nudiviruses (Table 2), which suggests that the baculoviruses and nudiviruses share a similar mode of transcription.
For the phylogenetic analysis (Figure 2C), the protein sequences of LEF-4, LEF-5, LEF-8, LEF-9 and P47 from the nudiviruses and selected baculoviruses were arranged sequentially and then aligned. In this tree, PmNV also formed a cluster with the nudiviruses. The tree showed a relatively large evolutionary distance between the baculo- virus cluster and the nudivirus cluster.
Genes involved in oral infectivity
PmNV010 (pif-5/odv-e56), PmNV015 (pif-2), PmNV039 (pif-1), PmNV072 (p74), PmNV088 (pif-6), PmNV093 (pif-3) and PmNV096 (pif-4) were homologous to genes
Yang et al. BMC Genomics 2014, 15:628 Page 4 of 24
http://www.biomedcentral.com/1471-2164/15/628
Table 2 Cross reference of the 37 conserved and 4 other baculovirus genes from AcMNPV with their homologs in the nudiviruses (OrNV, GbNV, HzNV-1 and HzNV-2) and PmNV
Function Gene name
Nudivirus
PmNV ORF/aa
Bracoviruses and hytrosaviruses
ORF (aa identity/similarity%) Accecssion no./ORF (aa identity/similarity%)
AcMNPV OrNV GbNV HzNV-1 HzNV-2 CcBV CiBV MdSGHV GpSGHV
37 baculovirus core genes
Replication dnapol 65 (8/24) 1 (12/30) 12 (16/36) 131 (24/46) 18 (24/46) 5/1091 NA NA 1 (8/22) 79 (9/24)
helicase 95 (9/26) 34 (12/31) 88 (11/28) 104 (16/31) 38 (16/31) 94/1271 NA NA - -
lef-1 14 - - - - - NA NA - -
lef-2 6 - - - - - NA NA - -
alk-exo 133 - - - - - NA NA - -
Transcription p47 40 (8/20) 20 (14/26) 69 (10/21) *75 (7/15) *63 (7/15) 14/419 CAR31573 (17/33) NA - -
lef-4 90 (8/21) 42 (15/31) 96 (14/33) 98 (10/19) 43 (10/19) 91/470 NA CAR40187 (6/14) - -
lef-5 99 (4/17) 52 (9/16) 85 (9/12) 101 (14/23) 40 (14/23) 52/175 CAT00573 (12/27) NA - -
lef-8 50 (9/24) 64 (17/34) 49 (18/35) 90 (17/33) 51 (17/33) 23/1032 CAR82252 (18/33) CBB83982 (3/6) - -
lef-9 62 (9/27) 96 (15/32) 24 (16/32) *75 (12/21) *63 (12/21) 58/574 NA NA - -
Oral infectivity pif-0 (p74) 138 (17/38) 126 (24/43) 45 (23/43) 11 (26/48) 106 (26/47) 72/684 CAR82260 (24/45) CAR40192(24/45) 39 (13/28) 1 (12/30)
pif-1 119 (11/25) 60 (20/34) 52 (18/32) 55 (22/38) 82 (22/38) 39/525 NA NA 29 (9/20) 102 (10/21)
pif-2 22 (14/27) 17 (20/35) 66 (20/34) 123 (25/43) 26 (25/43) 15/430 NA CAR40194 (17/32) 89 (12/26) 53 (20/34) pif-3 115 (14/23) 107 (22/38) 3 (24/40) 88 (28/43) 53 (28/43) 93/192 CAR82247 (20/38) NA 106 (14/23) 101 (5/12) pif-4 (19 k/odv-e28) 96 (14/31) 33 (20/35) 87 (17/31) 103 (9/16) 39 (9/16) 96/232 CAR31579 (17/36) CAR40196 (21/38) - -
pif-5 (odv-e56) 148 (10/24) 115 (14/32) 5 (16/34) 76 (22/42) 62 (21/42) 10/449 CAR31578 (15/33) CAR31577 (14/30) - 97 (11/25)
pif-6 (ac68) 68 (9/25) 72 (19/42) 55 (20/43) 74 (20/41) 64 (20/40) 88/143 CAR82241 (9/25) NA - -
Packaging, assembly, and release
38 k 98 (10/27) 87 (14/30) 1 (13/27) 10 (21/38) 108 (21/38) 59/282 CAR82239 (16/33) CAR40188 (19/38) - -
p6.9 100 - - - - - NA NA - -
**vlf-1 77 (6/18) 30 (6/15) 80 (12/26) 121 (10/22) 28 (10/22) 56/289 CAR40203 (16/35) CAR40190 (13/29) - -
**vlf-1 77 (5/18) 30 (4/10) 80 (9/26) 121 (8/22) 28 (8/22) 90/240 CAR40203 (19/35) CAR40190 (17/33) - -
vp39 89 15 64 89 52 - NA NA - -
vp1054 54 - - - - - NA NA - -
vp91/p95 83 (10/23) 106 (17/34 ) 2 (14/29) 46 (17/34) 89 (17/34) 9/675 NA NA - -
gp41 80 - - - - - NA NA - -
p6.9 100 - - - - - NA NA - -
p18 93 - - - - - NA NA - -
p33 92 (9/25) 113 (11/20) 7 (11/23) 13 (24/42) 104 (25/42) 8/228 NA NA - -
Yangetal.BMCGenomics2014,15:628Page5of24http://www.biomedcentral.com/1471-2164/15/628
Table 2 Cross reference of the 37 conserved and 4 other baculovirus genes from AcMNPV with their homologs in the nudiviruses (OrNV, GbNV, HzNV-1 and HzNV-2) and PmNV (Continued)
p40 101 - - - - - NA NA - -
p48 103 - - - - - NA NA - -
p49 142 - - - - - NA NA - -
odv-ec43 109 - - - - - NA NA - -
odv-e18 143 - - - - - NA NA - -
odv-e27 144 - - - - - NA NA - -
desmoplakin 66 - - - - - NA NA - -
ac53 53 - - - - - NA NA - -
Unknown function ac81 81 (10/21) 4 (23/41) 14 (20/41) 33 (32/54) 96 (33/54) 86/157 NA NA 108 (13/26) -
ac78 78 - - - - - NA NA - -
Other baculovirus genes
Transferase enzyme methyltransferase 69 (14/24) - - 37 (7/14) 93 (7/14) 6/401 NA NA - -
ODV envelope **odv-e66 46 (10/20) 12 (16/32) - - - 34/551 NA NA 47 (12/23) 5 (8/14)
**odv-e66 46 (12/24) 12 (15/30) - - - 36/635 NA NA 47 (13/28) 5 (9/16)
Anti-apoptosis iap 27 (6/12) 134 (7/11) 98 (7/14) 135 (14/23) 15 (14/23) 106/101 NA NA 78 (13/24) -
Occlusion body polyhedrin 8 (5/15) 16 (11/25) 65 (12/27) 68 (7/20) 70 (7/20) #20/423 NA NA 76(10/23) -
polyhedrin 8 - - - - ##1/452 - - - -
*In HzNV-1 both fused in the same ORF and appear to be fused into a single gene.
**The PmNV genome contains two ORFs that are homologous to vlf-1 and to odv-e66.
#Despite the [low] identity and similarity between PmNV020 and other polyhedrons from AcMNPV and the nudiviruses, this ORF does not in fact seem to express a functional polyhedrin.
##PmNV001 is a functional homolog of polyhedrin that shows no sequence homology to AcMNPV polyhedrin.
Bracoviruses and hytrosaviruses are included for comparison.
Yangetal.BMCGenomics2014,15:628Page6of24http://www.biomedcentral.com/1471-2164/15/628
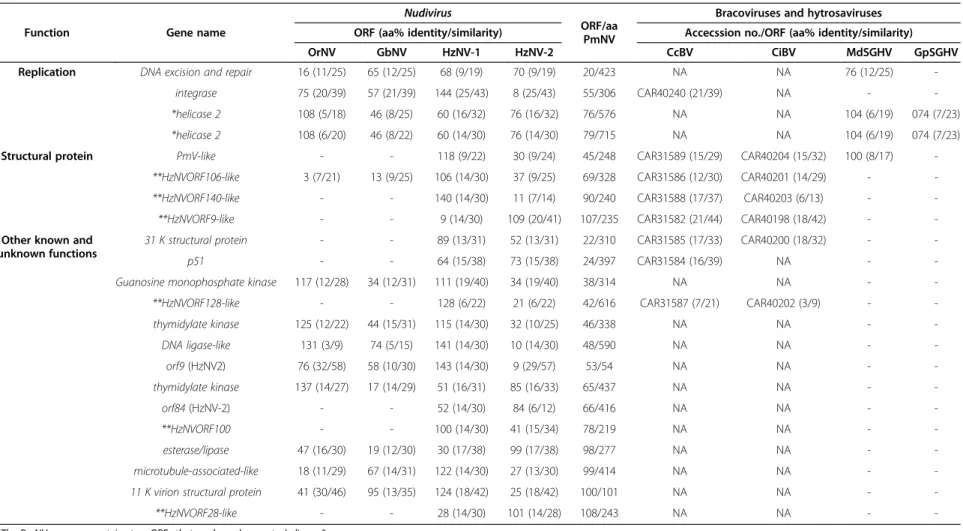
Table 3 Cross reference of PmNV genes that have homologs in the nudiviruses (OrNV, GbNV, HzNV-1 and HzNV-2), bracoviruses (CcBV and CiBV), hytrosaviruses (MdSGHV and GpSGHV)
Function Gene name
Nudivirus
ORF/aa PmNV
Bracoviruses and hytrosaviruses ORF (aa% identity/similarity) Accecssion no./ORF (aa% identity/similarity)
OrNV GbNV HzNV-1 HzNV-2 CcBV CiBV MdSGHV GpSGHV
Replication DNA excision and repair 16 (11/25) 65 (12/25) 68 (9/19) 70 (9/19) 20/423 NA NA 76 (12/25) -
integrase 75 (20/39) 57 (21/39) 144 (25/43) 8 (25/43) 55/306 CAR40240 (21/39) NA - -
*helicase 2 108 (5/18) 46 (8/25) 60 (16/32) 76 (16/32) 76/576 NA NA 104 (6/19) 074 (7/23)
*helicase 2 108 (6/20) 46 (8/22) 60 (14/30) 76 (14/30) 79/715 NA NA 104 (6/19) 074 (7/23)
Structural protein PmV-like - - 118 (9/22) 30 (9/24) 45/248 CAR31589 (15/29) CAR40204 (15/32) 100 (8/17) -
**HzNVORF106-like 3 (7/21) 13 (9/25) 106 (14/30) 37 (9/25) 69/328 CAR31586 (12/30) CAR40201 (14/29) - -
**HzNVORF140-like - - 140 (14/30) 11 (7/14) 90/240 CAR31588 (17/37) CAR40203 (6/13) - -
**HzNVORF9-like - - 9 (14/30) 109 (20/41) 107/235 CAR31582 (21/44) CAR40198 (18/42) - -
Other known and unknown functions
31 K structural protein - - 89 (13/31) 52 (13/31) 22/310 CAR31585 (17/33) CAR40200 (18/32) - -
p51 - - 64 (15/38) 73 (15/38) 24/397 CAR31584 (16/39) NA - -
Guanosine monophosphate kinase 117 (12/28) 34 (12/31) 111 (19/40) 34 (19/40) 38/314 NA NA - -
**HzNVORF128-like - - 128 (6/22) 21 (6/22) 42/616 CAR31587 (7/21) CAR40202 (3/9) - -
thymidylate kinase 125 (12/22) 44 (15/31) 115 (14/30) 32 (10/25) 46/338 NA NA - -
DNA ligase-like 131 (3/9) 74 (5/15) 141 (14/30) 10 (14/30) 48/590 NA NA - -
orf9 (HzNV2) 76 (32/58) 58 (10/30) 143 (14/30) 9 (29/57) 53/54 NA NA - -
thymidylate kinase 137 (14/27) 17 (14/29) 51 (16/31) 85 (16/33) 65/437 NA NA - -
orf84 (HzNV-2) - - 52 (14/30) 84 (6/12) 66/416 NA NA - -
**HzNVORF100 - - 100 (14/30) 41 (15/34) 78/219 NA NA - -
esterase/lipase 47 (16/30) 19 (12/30) 30 (17/38) 99 (17/38) 98/277 NA NA - -
microtubule-associated-like 18 (11/29) 67 (14/31) 122 (14/30) 27 (13/30) 99/414 NA NA - -
11 K virion structural protein 41 (30/46) 95 (13/35) 124 (18/42) 25 (18/42) 100/101 NA NA - -
**HzNVORF28-like - - 28 (14/30) 101 (14/28) 108/243 NA NA - -
*The PmNV genome contains two ORFs that are homologous to helicase 2.
**“HzNVORF” genes use the HzNV-1 ORF number.
Identity and similarity values in Tables2and3were calculated by ClustalX2 and GeneDoc and are different from the BLASTP results in Table4and in the text.
Yangetal.BMCGenomics2014,15:628Page7of24http://www.biomedcentral.com/1471-2164/15/628
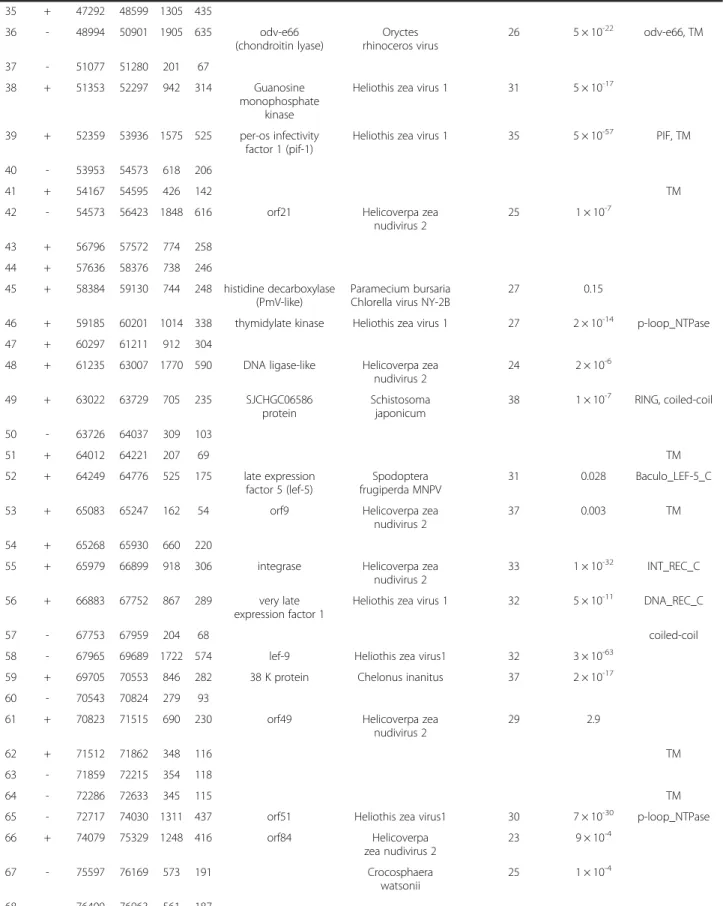
Table 4 Annotation of PmNV putative ORFs ORF Strand
Position Length Best BLAST match
Signature
Start End nt aa ORF, protein
encoded, or mass Species aa identity
(% match identity) E value
1 + 1 1359 1356 452 polyhedrin
2 - 1525 1989 462 154 coiled-coil
3 - 2033 4861 2826 942 von Willebrand
factor type A
Frankia sp. CN3 33 8 × 10-3 vWFA, ZF_RING_2
4 - 4954 5913 957 319
5 + 5998 9273 3273 1091 DNA polymerase Helicoverpa
zea nudivirus 2
26 2 × 10-102 POLBc
6 + 9354 10559 1203 401 methyltransferase Ceratitis capitata 33 FtsJ
7 - 10563 10772 207 69 TM
8 - 10779 11455 684 228 Ac92-like
(sulfhydryl oxidase)
Heliothis zea virus 1 42 1 × 10-17 Evr1_Alr, TM
9 - 11455 13482 2025 675 vp91 (capsid) Oryctes rhinoceros virus
31 7 × 10-32 ChtBD2, TM
10 + 13622 14971 1347 449 Odv-e56/pif-5 (envelope)
Heliothis zea virus 1 27 2 × 10-30 TM
11 - 13780 14331 549 183 TM
12 + 15057 15536 477 159
13 - 15325 15549 222 74 TM
14 + 15548 16807 1257 419 p47 Heliothis zea virus 1 24 1 × 10-5
15 - 16816 18108 1290 430 per-os infectivity factor 2 (pif-2)
Macrobrachium nudivirus
66 2 × 10-75 Baculo_44,
coiled-coil, TM
16 + 17191 17598 405 135 OHCU decarboxylase Sarcophilus harrisii 27 3 × 10-3 TM
17 - 18139 18987 846 282 HZV_115-like Oryctes rhinoceros virus
33 0.58 p-loop_NTPase,
NK superfamily
18 - 19071 20372 1299 433
19 + 20465 20869 402 134
20 + 20812 22083 1269 423 flap endonuclease 1 (FEN-1)
Heliothis zea virus 1 29 3 × 10-14
21 + 22080 22556 474 158
22 - 22553 23485 930 310 31 K structural protein Chelonus inanitus 24 1 × 10-9 23 + 23615 26713 3096 1032 RNA polymerase
(lef-8)
Heliothis zea virus 1 28 8 × 10-53 RNA_pol_Rpb2_6
24 + 26864 28057 1191 397 p51 late protein Heliothis zea virus 1 23 4 × 10-9 SBP_BACTERIAL_1
25 - 28160 31906 3744 1248 AF-4, ZnF_C2H2
26 + 31688 31912 222 74 TM
27 - 31969 32715 744 248
28 - 34005 34214 210 69
29 - 34463 42040 7572 2524 TM
30 - 42077 42649 570 190
31 + 43013 43204 189 63
32 - 43300 43512 210 70
33 + 43884 44783 897 299 tripartite
motif-containing protein 10-like
Alligator mississippiensis
37 7 × 10-9 RING, coiled-coil
34 - 44939 46594 1653 551 odv-e66
(chondroitin lyase)
Oryctes rhinoceros virus
29 1 × 10-23 odv-e66
Yang et al. BMC Genomics 2014, 15:628 Page 8 of 24
http://www.biomedcentral.com/1471-2164/15/628
Table 4 Annotation of PmNV putative ORFs (Continued)
35 + 47292 48599 1305 435
36 - 48994 50901 1905 635 odv-e66
(chondroitin lyase)
Oryctes rhinoceros virus
26 5 × 10-22 odv-e66, TM
37 - 51077 51280 201 67
38 + 51353 52297 942 314 Guanosine
monophosphate kinase
Heliothis zea virus 1 31 5 × 10-17
39 + 52359 53936 1575 525 per-os infectivity factor 1 (pif-1)
Heliothis zea virus 1 35 5 × 10-57 PIF, TM
40 - 53953 54573 618 206
41 + 54167 54595 426 142 TM
42 - 54573 56423 1848 616 orf21 Helicoverpa zea
nudivirus 2
25 1 × 10-7
43 + 56796 57572 774 258
44 + 57636 58376 738 246
45 + 58384 59130 744 248 histidine decarboxylase (PmV-like)
Paramecium bursaria Chlorella virus NY-2B
27 0.15
46 + 59185 60201 1014 338 thymidylate kinase Heliothis zea virus 1 27 2 × 10-14 p-loop_NTPase
47 + 60297 61211 912 304
48 + 61235 63007 1770 590 DNA ligase-like Helicoverpa zea nudivirus 2
24 2 × 10-6
49 + 63022 63729 705 235 SJCHGC06586
protein
Schistosoma japonicum
38 1 × 10-7 RING, coiled-coil
50 - 63726 64037 309 103
51 + 64012 64221 207 69 TM
52 + 64249 64776 525 175 late expression factor 5 (lef-5)
Spodoptera frugiperda MNPV
31 0.028 Baculo_LEF-5_C
53 + 65083 65247 162 54 orf9 Helicoverpa zea
nudivirus 2
37 0.003 TM
54 + 65268 65930 660 220
55 + 65979 66899 918 306 integrase Helicoverpa zea
nudivirus 2
33 1 × 10-32 INT_REC_C
56 + 66883 67752 867 289 very late
expression factor 1
Heliothis zea virus 1 32 5 × 10-11 DNA_REC_C
57 - 67753 67959 204 68 coiled-coil
58 - 67965 69689 1722 574 lef-9 Heliothis zea virus1 32 3 × 10-63
59 + 69705 70553 846 282 38 K protein Chelonus inanitus 37 2 × 10-17
60 - 70543 70824 279 93
61 + 70823 71515 690 230 orf49 Helicoverpa zea
nudivirus 2
29 2.9
62 + 71512 71862 348 116 TM
63 - 71859 72215 354 118
64 - 72286 72633 345 115 TM
65 - 72717 74030 1311 437 orf51 Heliothis zea virus1 30 7 × 10-30 p-loop_NTPase
66 + 74079 75329 1248 416 orf84 Helicoverpa
zea nudivirus 2
23 9 × 10-4
67 - 75597 76169 573 191 Crocosphaera
watsonii
25 1 × 10-4
68 - 76400 76963 561 187
Yang et al. BMC Genomics 2014, 15:628 Page 9 of 24
http://www.biomedcentral.com/1471-2164/15/628
Table 4 Annotation of PmNV putative ORFs (Continued) 69 - 76980 77966 984 328 HzNVORF106-like
protein
Cotesia congregata 24 1 × 10-6
70 - 77935 78555 618 206
71 - 78633 80174 1539 513
72 - 80247 82301 2052 684 p74 / pif-0
(envelope)
Heliothis zea virus 1 30 4 × 10-95 Baculo_74, TM
73 + 82393 83277 882 294 Zonadhesin Sorex araneus 49 2 × 10-52 TM
74 - 82986 83258 237 90 TM
75 + 83362 83934 570 190
76 + 84006 85736 1728 576 helicase 2 Danaus plexippus 30 1 × 10-46
77 + 85788 86687 897 299
78 - 86652 87311 657 219 orf100 Heliothis zea virus 1 23 0.014
79 + 87109 89256 2145 715 helicase 2 Danaus plexippus 28 5 × 10-39 TM
80 + 89240 92104 2862 954 coiled-coil
81 - 90949 91242 291 97
82 + 92108 93043 933 311
83 + 93036 93647 609 203
84 - 93644 95206 1560 520
85 - 95208 95618 408 136
86 + 95617 96090 471 157 Ac81-like Helicoverpa zea
nudivirus 2
36 3 × 10-21 TM
87 + 96083 97993 1908 636
88 + 98006 98437 429 143 Ac68-like/pif-6 Helicoverpa zea nudivirus 2
29 8 × 10-11 TM
89 - 98438 99691 1251 417
90 + 99781 100530 720 240 VLF-1 protein Helicoverpa zea nudivirus 2
28 5 × 10-5
91 - 100500 101912 1410 470 late expression factor 4
Helicoverpa zea nudivirus 2
26 4 × 10-18
92 - 101932 102255 321 107
93 - 102312 102890 576 192 per-os infectivity factor 3
Helicoverpa zea nudivirus 2
38 4 × 10-26 TM
94 - 102887 106702 3813 1271 helicase Helicoverpa zea
nudivirus 2
28 3 × 10-19 primase_Cte
95 + 106149 106370 219 73
96 + 106701 107399 696 232 19kda protein/
Odv-e28/pif-4
Oryctes rhinoceros virus
38 5 × 10-21 Baculo_19, TM
97 - 107374 108123 747 249
98 + 108113 108946 831 277 Esterase/lipase Helicoverpa zea nudivirus 2
27 3 × 10-4 AAA_17, TM
99 - 108943 110187 1242 414 GrBNV_gp67-like Oryctes
rhinoceros virus
29 9 × 10-8 EF_HAND_1,
coiled-coil 100 - 110290 110595 303 101 11 K virion
structural protein
Oryctes rhinoceros virus
32 4 × 10-6 TONB_DEPENDENT_
REC_1, TM 101 - 110705 111106 399 133
102 + 111081 111881 798 266 103 - 111883 113868 1983 661 104 + 113631 113864 231 77 105 + 114593 114865 270 90
Yang et al. BMC Genomics 2014, 15:628 Page 10 of 24
http://www.biomedcentral.com/1471-2164/15/628
involved in per os infection. These seven genes are con- served across the baculoviruses and nudiviruses, and they are all essential for the occlusion derived virus (ODV) infectivity of the baculoviruses, either by specific binding to the midgut cells (P74, PIF-1 and PIF-2) [22,23]
or by a mechanism that has not yet been determined (PIF- 4, PIF-5 and PIF-6) [24-26]. Although PIF-3 was shown not to be involved in specific attachment and fusion [23], deletion of its N-terminal nuclear localization signal prevents per os infectivity [27].
The shared importance of these per os infectivity fac- tors implies that despite the absence of OBs in the nudi- viruses, these viruses nevertheless all share a very similar oral infection mechanism. Moreover, four homologous genes that have relatively high identity and similarity (p74, pif-1, pif-2 and pif-3) are also found in the hytrosa- viruses, indicating that the baculoviruses, nudiviruses and hytrosaviruses all have the same important ancestral conserved model for virus infectivity. Interestingly, BLASTP search showed that PmNV015 (pif-2) most closely matched Macrobrachium nudivirus (MRNuV) PIF-2 (Accession No. : AFP33714) with an identity of 66%. To date, only one other MRNuV gene has been se- quenced (IAP), but we note that unlike the other insect nudiviruses, both PmNV and MRNuV are aquatic vi- ruses, and it is possible that these two viruses - perhaps together with Penaeus vannamei single nucleopolyhe- drovirus, PvSNPV - are quite closely related.
The constructed phylogenic tree of the oral infectivity genes p74, pif-1, pif-2 and pif-3 (Figure 2D) is similar to the DNA polymerase and Helicase trees, providing fur- ther evidence that PmNV is more closely related to the nudiviruses than the baculoviruses.
Proteins related to viral packaging, assembly, and release There are 18 baculovirus core genes involved in viral structure formation, but only four of these genes have
homologs that could be found in the PmNV genome:
p33(PmNV008), vp91 (PmNV009), vlf-1 (PmNV056 and PmNV090) and 38 k (PmNV059) (Table 2). The PmNV genome even lacks a homolog to vp39, which is found in the nudiviruses and is thought to encode the major cap- sid protein [28].
P33 is a component of the occluded virions of AcMNPV, CuniNPV (Culex nigripalpus nucleopolyhedro- virus) and HzSNPV (Helicoverpa zea single-nucleocapsid nucleopolyhedrovirus) [29-31]. AcMNPV P33 has been shown to interact with the human tumor suppressor p53 to enhance p53-induced apoptosis [32]. However, while AcMNPV P33 also interacts with Spodoptera frugiperda P53 (SfP53) and oxidizes SfP53 in vitro, it does not enhance SfP53-mediated apoptosis in Sf9 cells [33].
AcMNPV P33 has also been demonstrated to have sulfhy- dryl oxidase activity [34] and it is required for budded virus production and for the formation of multiply en- veloped occlusion-derived nucleocapsids [35]. VP91 is expressed at the late stage of viral infection and is present in the capsid structure of OpMNPV (Orgyia pseudotsu- gata multicapsid nucleopolyhedrovirus) [36]. VLF-1 is expressed at the very late stage and is involved in the pro- duction of the AcMNPV nucleocapsid [37]. Gene 38 K is essential for budded virus formation and the nucleocapsid assembly of AcMNPV [38]. Taken together, the low num- ber of homologs in this functional group indicates that the baculoviruses and nudiviruses are highly divergent in terms of their viral structural proteins.
Proteins of unknown function
The best match to PmNV086 was the Ac81-like protein of HzNV-1 and HzNV-2 (BLASTP: E = 3 × 10-21) (Table 4). In BmNPV (Bombyx mori nucleopolyhedro- virus), the Ac81 homolog (ORF67) is a non-structural protein of unknown function that interacts with host Table 4 Annotation of PmNV putative ORFs (Continued)
106 - 114757 115062 303 101 apoptosis
inhibitor IAP-1
Buzura suppressaria NPV
43 2 × 10-13 BIR
107 - 115114 115821 705 235 HzNVORF9-like Chelonus inanitus 25 4 × 10-10
108 + 115917 116648 729 243 orf101 Helicoverpa zea
nudivirus 2
23 0.1 ribokinase_
pfkB_like, TM 109 - 116750 116896 144 48
110 - 116971 117372 399 133 TM
111 + 117179 117469 288 96
112 + 117529 118617 1086 362 TM
113 + 118674 118928 252 84 sortilin-related receptor precursor
Acyrthosiphon pisum 42 9 × 10-9 LDLRA_2
114 + 118723 118911 186 62 TM
115 + 119035 119493 456 152 TM
Yang et al. BMC Genomics 2014, 15:628 Page 11 of 24
http://www.biomedcentral.com/1471-2164/15/628
actin [39]. The protein sequence encoded by PmNV086 was predicted to have a transmembrane domain by SMART, and Pfam found a partial match (E = 0.16) in the eukaryotic RHD3 protein (Root hair defective 3 GTP- binding protein).
Non-core AcMNPV genes
We found four other putative PmNV ORFs homologous to baculovirus genes that are not included in the 37 core genes: PmNV006 (methyltransferase), PmNV034 (odv-e66), PmNV036 (odv-e66), PmNV106 (iap) (Table 2).
Methyltransferase is expressed in the late phase of infection and has cap O-dependent methytransferase activity [40]. However, deletion studies with methyltrans-
ferase-defective AcMNPV have shown that it is not essen- tial for the budded virus or for occluded virus production, and that it has no effect on AcMNPV [41].
The PmNV genome has two neighboring ORFs with the same orientation (PmNV034 and PmNV036) that are homologous to odv-e66. ODV-E66 is a late expression, structural component of occluded-derived virus (ODV) envelopes [42]. The N-terminus of ODV-E66 contains a highly hydrophobic INM-sorting (inner nuclear mem- brane-sorting) motif that traffics AcMNPV viral proteins to enhance viral assembly [43]. However, despite their homology, PmNV034 and PmNV036 have different lengths, relatively low identity and similarity (38% and 53%, respectively), and the hydrophobic INM-sorting
Figure 2 Phylogenetic analysis of selected DNA viruses and PmNV. Multiple alignments of (A) DNA polymerase, (B) Helicase, (C) LEF-4, LEF-5, LEF-8, LEF-9 and P47 (D) P74, PIF-1, PIF-2 and PIF-3 protein sequences were performed using BioEdit. The trees were inferred using MEGA5.2 and the neighbor-joining method. The robustness of each tree was tested using bootstrap (1000) analysis. The percent values are indicated at the nodes. Organisms included in this analysis, with abbreviated names: Autographa californica multiple nucleopolyhedrovirus (AcMNPV), Bombyx mori nucleopolyhedrovirus (BmNPV), Lymantria xylin nucleopolyhedrovirus (LyxyMNPV), Cydia prmonella granulovirus (CpGV), Agrotis segetum granulovirus (AgseGV), Spodoptera litura granulovirus (SpliGV), Neodiprion sertifer nucleopolyhedrovirus (NeseNPV), Neodiprion lecontei nucleopolyhedrovirus (NeleNPV), Neodiprion Abietis nucleopolyhedrovirus (NeabNPV), Culex nigripalpus nucleopolyhedrovirus (CuniNPV), Heliothis zea virus 1 (HzNV-1), Helicoverpa zea nudivirus 2 (HzNV-2), Oryctes rhinoceros nudivirus (OrNV), Gryllus bimaculatus nudivirus (GbNV), Musca domestica salivary gland hypertrophy virus (MdSGHV), Glossina pallidipes salivary gland hypertrophy virus (MdSGHV), African swine fever virus (ASFV). Accession numbers of all the sequences are listed in Additional file 8: Table S6.
Yang et al. BMC Genomics 2014, 15:628 Page 12 of 24
http://www.biomedcentral.com/1471-2164/15/628
sequence was found only in PmNV036 and not in PmNV034. AcMNPV ODV-E66 has also recently been shown to act as a chondroitin lyase [44]. Interestingly, both PmNV034 and PmNV036 contain all five of the conserved catalytic residues that are found in AcMNPV ODV-E66, suggesting that either or both of these pro- teins might function as a chondroitin lyase.
PmNV106 contains a single predicted BIR (baculoviral inhibitor of apoptosis repeat) domain. This domain acts directly on the caspase family of protease enzymes to block apoptosis and thereby allow the virus more time to replicate.
The IAP (inhibitor of apoptosis) family of proteins contains 1 ~ 3 BIR domains in the N terminal and an optional RING domain in the C terminal. PmNV106, which lacks a pre- dicted RING domain and is shorter than most other IAPs (Additional file 8: Table S6 and Additional file 9: Figure S3), shares an identity of less than 60% with other IAPs.
The PmNV genome contains two ORFs with some homology to polyhedrin, PmNV001 and PmNV020. LC- MS analysis has shown that the polyhedrin protein purified from PmNV occlusion bodies is derived from PmNV001 [19]. Meanwhile, although three nudivirus ORFs (Hz1V068, OrNV ORF16 and GbNV ORF65) were previously reported to show some homology to AcMNPV polyhedrin [18], and our BLASTP search also found a fourth match in Hz2V070, there is no functional evidence that polyhe- drin protein is actually expressed by any of these ORFs.
The evolutionary relationship between these low identity, possible polyhedrin nudivirus homologs and baculoviral polyhedrin still remains unclear, but we tentatively con- clude that PmNV020 is unlikely to be a polyhedrin gene.
PmNV polyhedrin is discussed in more detail in the second half of this study.
PmNV putative ORFs that are homologous to nudivirus, bracovirus and hytrosavirus genes
Enzymes involved in DNA replication
The PmNV genome contains four putative ORFs, PmNV 020 (DNA excision and repair) PmNV055 (integrase), PmNV076 (helicase2) and PmNV079 (helicase2), that are homologous to genes involved in DNA replication.
PmNV020, in addition to being a homolog to four nudi- virus ORFs that are homologous to AcMNPV polyhedrin, has a predicted 29% amino acid identity with Hz1V068 and Hz2V070 (BLASTP: E = 3 × 10-14). A Pfam database search revealed that the N-terminus of all three proteins matched the XPG (Xeroderma pigmentosum complemen- tation group G) N-terminal domain with E-values of around 0.03 (PmNV020 33-110 aa = 0.03, Hz1V068 = 0.027 and Hz2V070 = 0.028). The presence of this domain in Hz2V070 led Burand et al. [7] to propose that it is a DNA excision and repair enzyme that restores damaged DNA. Homologs of this enzyme are found among the nudiviruses and in the Musca domestica salivary gland
hypertrophy virus (MdSGHV), but not in the baculovirus genomes. The baculoviruses encode various other proteins for DNA repair instead. For example, some of the nucleo- polyhedroviruses use photolyase to repair UV-damaged DNA [45]. PmNV055 (integrase) most closely matched the integrase of HzNV-1 (BLASTP: E = 1 × 10-32) (Table 4), which phylogenetic analysis suggests was formed by duplication of the baculovirus and nudivirus conserved vlf-1 gene [46]. PmNV055 is predicted to belong to the phage integrase family (Pfam: PmNV055 105-278 aa, E = 7.8 × 10-16). Members of this family mediate unidirectional site-specific recombination between two DNA recognition sequences [47]. Tyrosine is the key catalytic site of this enzyme activity, and a catalytic tyrosine was found on one of the six possible active sites of PmNV055 predicted by Pfam. Integrase mediates the excision and integration of viral genetic material into the host genome. This process is essential for Microplitis demolitorbracovirus (MdBV) [48], and HzNV-1 also has a similar mechanism that integrates viral DNA into the host chromosome for persistent infection [49]. However, it is still unclear if this protein has any functional role in PmNV.
BLASTP found that PmNV076 and PmNV079 both matched the same Danaus plexippus protein with an identity of 30% (E = 1 × 10-46) and 28% (E = 5 × 10-39), re- spectively (Table 4). The same two ORFs also matched HzNV-1 putative helicase 2 with 27% identity (BLASTP:
E = 1 × 10-39 and 7 × 10-35, respectively). However, these two ORFs have relatively low amino acid sequence identity and similarity (17% and 35%, respectively), which means that if they really are homologs to helicase 2, then the original gene duplication must have occurred a long time ago. The genomes of the other sequenced nudi- viruses each contain only a single instance of a gene that is homologous to helicase 2.
Structural proteins
Four putative ORFs, PmNV045 (PmV-like), PmNV069 (HzNVORF106-like), PmNV090 (HzNVORF140-like) and PmNV107 (HzNVORF9-like), were homologous to bra- covirus proteins. Proteomic analysis showed that these proteins were associated with the bracovirus particles [50], suggesting that the PmNV proteins might also be structural proteins involved in viral packaging, assembly, or release.
Proteins with other known and unknown functions
Fourteen other PmNV ORFs are homologous to nudi- virus and bracovirus genes/ORFs with miscellaneous or unknown functions.
PmNV024 most closely matched the HzNV-1 p51 gene (BLASTP: E = 4 × 10-9) (Table 4), which is a late
Yang et al. BMC Genomics 2014, 15:628 Page 13 of 24
http://www.biomedcentral.com/1471-2164/15/628