1
Grouper (Epinephelus coioides) CXCR4 is expressed in response to pathogens infection 1
and early stage of development 2
3
Ching-Yu Lina, Young-Mao Chena,b,c, Hao-Hsuan Hsua,c, Chia-Tai Shiua,c, Hsiao-Che 4
Kuoa,b,c, Tzong-Yueh Chena,b,c* 5
6
aLaboratory of Molecular Genetics, Institute of Biotechnology, College of Bioscience and 7
Biotechnology, National Cheng Kung University, Tainan 70101, Taiwan 8
bResearch Center of Ocean Environment and Technology, National Cheng Kung University, 9
Tainan 70101, Taiwan 10
cAgriculture Biotechnology Research Center, National Cheng Kung University, Tainan 11 70101, Taiwan 12 13 *Corresponding author: 14 Dr. Tzong-Yueh Chen 15
Laboratory of Molecular Genetics, Institute of Biotechnology, College of Bioscience and 16
Biotechnology, National Cheng Kung University, Tainan 70101, Taiwan 17
Telephone: 886-6-2757575 ext. 65622 ext.610 18
Fax: 886-6-2766505 19
E-mail: ibcty@mail.ncku.edu.tw 20
Source:Developmental and Comparative Immunology, Vol. 36, No. 1, pp. 112-120 Year of Publication:2011
ISSN:0145-305X Publisher:Elsevier
DOI:10.1016/j.dci.2011.06.009 © 2011 Elsevier Ltd. All rights reserved
2 Abstract
21
Chemokine (C-X-C motif) receptor 4 (CXCR4) from orange-spotted grouper (Epinephelus 22
coioides) was identified and characterized in this study. gCXCR4 shared common features 23
in protein sequence and predicted structure of CXCR4 family. This suggested that gCXCR4 24
is a member of G protein-coupled receptors with seven transmembrane domains. The 25
expression patterns revealed that gCXCR4 may play a key role in early development of 26
grouper. Furthermore, overexpression of gCXCR4-GFP for 48 h had significant effects on 27
the GF-1 cell viability. gCXCR4 protein was mainly expressed in the marginal zone of head 28
kidney and on the surface of intestinal villi. gCXCR4 expression can be detected in all the 29
examined tissues and significantly up-regulated in eye and brain, which are the main targets 30
for nervous necrosis virus (NNV) infection and replication. gCXCR4 gene expression can 31
be induced in the spleen and eye by lipopolysaccharide and NNV, respectively. Our data 32
suggested that gCXCR4 may not only play a role in the early immune response to microbial 33
infection but also restrain to the immune system and central nervous system. 34
Keywords: Epinephelus coioides; CXCR4; G-protein-coupled receptors; chemokine 35
3 1. Introduction
37
The orange-spotted grouper, Epinephelus coioides, is a commercially important fish that is 38
widely farmed in tropical waters of many countries. Considerable economic losses have 39
been sustained in grouper aquaculture due to the infection of grouper by piscine nodavirus, 40
i.e. nervous necrosis virus (NNV). The virus causes viral nervous necrosis (VNN) on 41
grouper hatchery larvae and juvenilies, resulting in a high mortality rate (80-100%) (Kuo et 42
al., 2011; Munday et al., 2002). 43
Chemokines are a group of small molecular weight (6–14 kDa) cytokines which play 44
important roles not only in against microbial infection by guiding leukocyte migration but 45
also in embryonic growth and development (Kim et al., 1999; Olson et al., 2002). 46
Chemokines can be classified into four different kinds, CXC, CC, C and CX3C (Murphy et 47
al., 2000), according to its cysteine motif in the N-terminal region. Chemokine receptors are 48
a group of G protein-coupled receptors with seven transmembrane domains. Upon 49
stimulation by chemokines, chemokine receptors trigger a series of intracellular signal 50
transductions via interaction with the G proteins. To this day, different chemokine receptors 51
have been found on various cells such as monocytes, T lymphocytes, B lymphocytes, 52
natural killer cells, macrophages, endothelial cells and neuron cells in mammals (Horuk et 53
al., 2009). To date, CXCR4 has been identified in several species including human 54
(Federsppiel et al., 1993), mouse (Heesen et al., 1996) and dog (Tsuchida et al., 2007), but 55
4
less is known in fish (Daniels et al., 1999; Chong et al., 2001; Alabyev et al., 2000; Jia and 56
Zhang, 2009), and no functional characterization of CXCR4 in orange-spotted grouper has 57
been reported. 58
Chemokines and their receptors serve an important role in viral infections and among 59
the chemokine receptors, CXCR4 is also a co-receptor for human immunodeficiency virus 60
(HIV) entry into target cells (Feng et al., 1996; Berson et al., 1996). Nonetheless, CXCR4 is 61
not only involved in the pathogenesis of viral infections but also plays a critical role in 62
organogenesis and embryonic development-related vascularization, lymphopoiesis and 63
myelopoiesis (Tachibana et al., 1998). CXCR4 deficiency produces a lethal phenotype and 64
abnormal development of central nervous system, such as abnormal migration of granule 65
cells and an altered location of Purkinje cells in mice malformed cerebellum (Ma et al., 66
1998). In fish, CXCR4 has been found in the early stage of zebrafish embryo, it can be 67
detected in the lateral mesoderm and posterior midbrain (Chong et al., 2001), moreover the 68
migration of lateral-line-primordium is impeded in CXCR4 homologue-mutated zebrafish 69
and is completely absent in SDF-1a defective zebrafish (Valentin et al., 2007). In addition, 70
CXCR4 plays a crucial role for tissue polarity (Haas and Gilmour, 2006). CXCR4 71
homologue deficiency leads to the random migration of cells and the loss of coordinated 72
motility within the posterior lateral line primordium in zebrafish (Haas and Gilmour, 2006). 73
These observations indicate that CXCR4 is multifunctional and plays crucial roles in 74
5 embryonic growth development and hematopoiesis. 75
However, no functional characterization of grouper CXCR4 has been reported. 76
Previously, a partial portion of grouper CXCR4 cDNA was identified by subtractive cDNA 77
hybridization from healthy and NNV-infected groupers (Chen et al., 2010). In the present 78
study, orange-spotted grouper CXCR4 (gCXCR4) was cloned and the expression profile 79
that response to lipopolysaccharide (LPS) and NNV infection was investigated. In addition, 80
we showed that cell proliferation was impeded after gCXCR4 overexpression for 48 h. 81
6 2. Materials and methods
83
2.1. Fish and challenge experiments 84
Fish weighing approximately 300g (150 days post-hatching) and different ages (1-40 85
days-old) of orange-spotted grouper (E. coioides) were collected from an indoor fish farm in 86
Linyuan and maintained in 10L containers at 27 ± 1 °C. For challenge experiments, 15 87
300g-in-weight-fish were divided into five groups (n=3 per group) and challenged with 100 88
μl phosphate buffered saline (PBS) contained approximately 20 μg of purified Escherichia 89
coli 0127:B8 LPS (Sigma-Aldrich, St. Louis, MO) per fish via intraperitoneal injection. 90
Fish with 100 μl PBS injection was used as a control group. The fish were sacrificed and the 91
spleens were collected at 0, 6, 24, 48 and 72 h post-injection. In the experiments of virus 92
challenging, juvenile groupers (about 0.8 g in weight, 40–45 days post-hatching) were 93
collected from Linyuan fish farms in southern Taiwan. Twelve juvenile groupers were 94
divided into two groups, NNV infection group and control group. Each six fish were 95
immersed into 500 ml of rearing water which contained 50 ml of a viral solution (106 96
TCID50/0.1 ml) or saline for 2 h. The fish were transferred to a virus-free aquarium, which 97
had been exposure to ultraviolet (UV) light for 24 h, and cultured at 28 °C. Real time PCR 98
was then used to confirm the fish was infected by NNV after 72 h of challenging. 99
2.2. Total RNA extraction and cDNA synthesis 100
Eye or whole fish samples (n=3 per group) were used for total RNA extraction by 101
7
homogenized using a MagNALysis homogenizer (Roche, Basel, Switzerland) following the 102
manufacture’s recommendations of TRIzol™ (Invitrogen) method.. cDNA was synthesized 103
with 2 μg RNA, 0.1 μM oligo dT primer, 12.5 μM dNTP (Bioman Scientific Co. Ltd., 104
Taipei, Taiwan) and 50 units Molony Murine Leukemia Virus (MMLV) reverse transcriptase 105
(Promega, Madison, WI) at 42 oC for 1 h. 106
RNA and cDNA were quantified using an Ultrospec 3300 Pro spectrophotometer 107
(Amersham Biosciences, Piscataway, NJ, USA); nucleic acids were diluted using sheared 108
salmon sperm DNA (5 ng mL-1) as a carrier. 109
2.3 RNA gel electrophoresis 110
To confirm the integrity of RNA samples, the extracted RNA was evaluated by RNA 111
electrophoresis. In brief, 2 μl RNA sample was mixed with 18 μl 1× reaction buffer (1× 112
MOPS, 20 % formaldehyde and 50 % formamide), 2 µl of 10× formaldehyde gel loading 113
buffer (50 % glycerol, 10 mM EDTA, pH8, 0.25% bromphenol blue and 0.25 % xylene 114
cyanol) and was visualized by using ethidium bromide staining 115
2.4. Rapid amplification of cDNA ends (RACE) 116
A 950 bp cDNA fragment obtained from our previous study by subtractive cDNA 117
hybridization from healthy and NNV infected groupers (Chen et al., 2010). The sequence 118
showed 75% similarity to CXCR4 from Psetta maxima by blasting 119
(http://www.ebi.ac.uk/blastall/).Full length cDNA was obtained by 5’/3’ RACE which was 120
8
performed by using 5’/3’ RACE Kit. Gene-specific primers (Table 1) for 5’ and 3’ RACE 121
were designed based on partial sequence of gCXCR4 For 5’ RACE, mRNA was transcribed 122
by MMLV reverse transcriptase (Sigma-Aldrich) with primer gCXCR4-5SP1and 123
gCXCR4-5SP2 and gCXCR4-5SP3 were used for PCR and nested PCR. 3’ RACE was 124
performed by using primers gCXCR4-3SP1 and gCXCR4-3SP2 for PCR and nested PCR, 125
respectively. PCR condition was one cycle of 3 min at 95 oC, followed by 35 cycles each at 126
95 oC for 30 s, 58 oC for 30 s, 72 oC for 30s and a final extension at 72 oC for 10 min. The 127
primers gCXCR-F and gCXCR-R were used to amplify the gCXCR4 cDNA fragment. 128
2.5 Bioinformatic analyses of gCXCR4 129
The transmembrane domains, extracellular domains and cytoplasmic domains of CXCR4 130
were identified by the TMHMM Server 2.0 program 131
(http://www.cbs.dtu.dk/services/TMHMM/). The protein sequences of different CXCR4 132
species were obtained from GenBank and were aligned using Vector NTI 10 software. A 133
phylogenetic tree of CXCR4 was constructed by the neighbor-joining method using 134
MEGA4.0. The reliability of the tree was established by bootstrap analysis, based on 1,000 135
bootstrap replications. 136
2.6. RT-PCR 137
The tissue distribution of gCXCR4 gene expressions was investigated by RT-PCR. Total 138
RNA was extracted from different tissues of grouper such as eye, fin, gill, muscle, head 139
9
kidney, heart, spleen, intestine and brain. PCR condition was one cycle of 3 min at 95 oC, 140
followed by 35 cycles each at 95 oC for 30 s, 58 oC for 30 s, 72 oC for 30 s and a final 141
extension at 72 oC for 10 min. To detect the expression of gCXCR4 in different 142
developmental stages of grouper larvae, total RNA were prepared from pooled larvae that 143
contained 20 fish fry in each group at 1, 2, 4, 6, 8, 10, 14, 18 and 20-days post hatch (dph), 144
and 3 fish larvae were pooled at 24, 26, 28, 30, 32, 34, 38 and 40 dph. 145
2.6. Real Time-quantitative PCR 146
Real-time quantitative PCR was performed by StepOne™ real-time PCR system (Applied 147
Biosystems, Foster City, CA, USA). 1ul of cDNA (from 100 ng RNA) was mixed with 12.5 148
μl 2×SYBR® Green Master Mix (Applied Biosystems) and 1 μl of each 10 μM specific 149
primer (Table 1). The thermal profile for real-time PCR was 95 °C for 10 min followed by 150
40 cycles of 95 °C for 30 s, 60 °C for 30 s and a final stage at 95 °C for 15 s, 60 °C for 1 151
min, 95°C for 15 s. The results of real-time PCR were analyzed with StepOne Software 152
v2.1. 153
2.7 Statistical analysis 154
The CT for gCXCR4 and β-actin were determined for each sample. β-actin was used as 155
internal control. △CT (Differences between gCXCR4 and β-actin) was calculated to 156
normalize the differences in the efficiency of reverse transcription reactions. The △CT for 157
each sample was subtracted from the △CT of the calibrator, and the difference was 158
10
designated as the △△CT value. The relative expression level of gCXCR4 could be 159
calculated by 2-△△CT. All real-time PCR data were subjected to analysis of t-test and are 160
presented as the mean ± S.E. of the relative mRNA expression. P-values of < 0.05 were 161
considered significantly different. 162
2.8 Plasmid construction 163
To prepare anti-gCXCR4 antiserum, extracellular domain I and III of gCXCR4 were 164
constructed into pET29b expression vector (Novagen, USA) by PCR using primers. Primers 165
gCSCR4-EXI-F (BamHI) and gCSCR4-EXI-R (SalI) were used for extracellular domain I 166
and gCXCR4-EXIII-F (SalI) and gCXCR4-EXIII-R (XhoI) were used for amplifying 167
extracellular domain III (181-215 a.a.). This recombinant plasmid was named 168
pET29b-gCXCR4-EXI-EXIII which can express a fusion protein of gCXCR4 extracellular 169
domains I and III along with a 6×His tag. 170
The gCXCR4 overexpression vector, pcDNA3.1-gCXCR4-GFP, was constructed by 171
PCR amplifying gCXCR4 using primers gCXCR-F and gCXCR4-GFP-R (Table 1) and the 172
PCR products were then cloned into the pcDNA3.1-CT-GFP-TOPO expression vector 173
(Invitrogen). The inserted DNA fragments of each clone were confirmed by sequencing 174
(Mission Biotech Co., Ltd., Taipei, Taiwan). 175
2.9 Recombinant protein and anti-gCXCR4 antiserum preparation 176
The gCXCR4-EXI-EXIII-His recombinant protein was expressed by transforming 177
11
pET-29b-gCXCR4-EXI-EXIII intoBL21(DE3) cells (Novagen)and induced by adding 178
isopropyl-beta-D-thiogalactopyranoside (IPTG; MDBIO, Frederick, MD) to a final 179
concentration of 0.1 mM. Protein purification was performed using a HisTrap HP 1 ml 180
column (Amersham Biosciences, Piscataway, NJ). Antisera against gCXCR4 was obtained 181
by immunizing (injection of 1 mg/ml protein which was mixed with Freund's complete 182
adjuvant [Sigma-Aldrich] on days 1, 14 and 28) a New Zealand White rabbit (Taiwan 183
Livestock Research Institute, Tainan, Taiwan) with recombinant gCXCR4-extracellular 184
domain I-extracellular domain III fusion protein. The antiserum collected at day 0 (before 185
treatment) was used as control. The blood samples were incubated at 37℃for 1 h and left 186
overnight at 4 ℃ . The supernatant (containing rabbit anti-gCXCR4 antiserum) was 187
collected after centrifuging at 900 ×g for 10 min at 4 °C. The rabbit anti-gCXCR4 188
antiserum was stored at –20 °C. 189
2.10 Immunofluorescence staining 190
The head kidney and intestines were obtained from healthy groupers and treated with 30 % 191
sucrose at 4 ℃ overnight. The different tissue blocks were covered with an optimal cutting 192
temperature compound (Tissue-Tek®; Sakura Finetek, Tokyo, Japan), and the samples were 193
slowly placed into liquid nitrogen. The frozen tissue block was transferred into a cryotome 194
cryostat and 5 μm-thick sections were cut. Each slide was fixed with 3% paraformaldehyde 195
(Kanto Chemical, Tokyo, Japan) and incubated at room temperature for 30 min. The 196
12
samples were then washed with 1× PBST (0.1% Tween 20, 1× PBS) and blocked with 5 % 197
skim milk. Rabbit anti-gCXCR4 antisera (1:200 dilution) were added and subsequence 198
Alexa Fluor® 594 goat anti-rabbit IgG (1:200 dilution) (H+L) (Invitrogen) secondary 199
antibody was added. The nuclei were stained with Hoechst 33342 (Invitrogen) at room 200
temperature for 20 min, then washed extensively with 1×PBS and mounted on a coverslip 201
with mounting medium. 202
2.11 Cell proliferation assay 203
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrasodium bromide (MTT) assay was performed 204
using Cell Proliferation Kit I ( Roche ) to analyze the effects of gCXCR4-GFP 205
overexpression. 5×104 Epinephelus coicoides fin cells (GF-1, BCRC 960094) were seeded 206
in 24-well plate and grown in a humidified incubator operating at 28°C in an antibiotic-free 207
L15 medium (Life Technologies, Carlsbad, CA, USA) supplemented with 5% v/v 208
heat-inactivated fetal bovine serum (FBS) (Chen et al., 2008). After 24hr, when cells 209
attached completely, pcDNA3.1-CT-GFP-CXCR4 or pcDNA3.1-CT-GFP vector was 210
transfected using Lipofectamine 2000 (Invitrogen). Then 20μl of MTT labeling reagent was 211
added to each well and cultured for 4 hours in incubator. To dissolve formazan crystals, 212
200μl solubilization solution was added to each well. Covered with tinfoil and agitate cells 213
on orbital shaker for 10 min. Read absorbance at 570 nm. 214
13 3. Results
215
3.1. Characterization of full-length gCXCR4 216
Orange-spotted grouper CXCR4 (gCXCR4) contained an open reading frame (ORF) of 217
1,104 nucleotides encoding a 367 a.a. protein with a predicted molecular weight of 40.37 218
kDa (Fig. 1). The structure of the protein was predicted to have seven-transmembrane 219
domains, four extracellular domains and four cytoplasmic domains. The DRY motif was 220
found at the second intracellular loop. A conserved cysteine residue on each of the four 221
extracellular domains located at positions Cys33, Cys118, Cys198 and Cys292. The C-terminal 222
is a region rich in serine and threonine residues. 223
The results of different species CXCR4 alignment (Fig.2) showed that gCXCR4 not 224
only similar to fish (zebrafish [58%], carp [54%], rainbow trout [53%] and turbot [52%]) 225
but also to mammalian (human [51%) and mouse [50%)) and the conserved regions 226
appeared in seven putative transmembrane domains (Fig. 2). The sequence of extracellular 227
domains showed very diverse in different species. gCXCR4 has 22.1%, 8.5% , 28.8%, 228
32.2%, 33.6% and 29.3% similarity to human, mice, common carp, zebrafish, rainbow trout 229
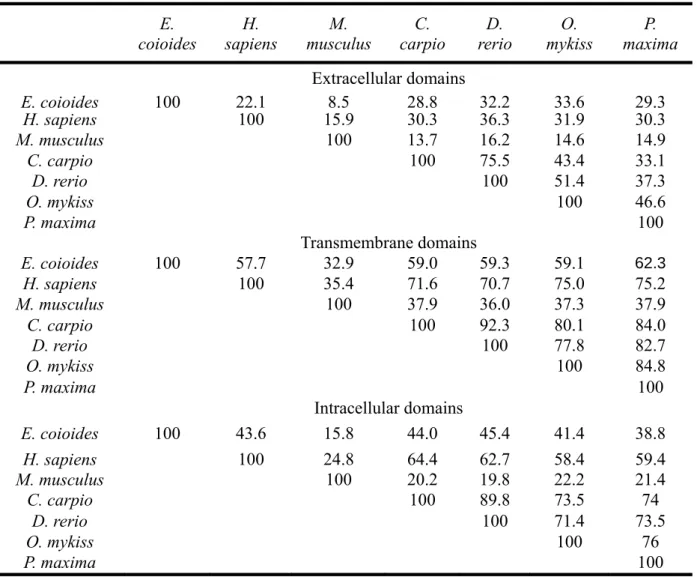
and turbot, respectively (Table 2). 230
3.2. Phylogenetic analysis of gCXCR4 231
There are six groups of CXCRs on the phylogenetic tree. Interestingly, within the group of 232
CXCR4, mammalian and fish origins of CXCR4s was clear separated except for gCXCR4 233
14
and Petromyzon marinus CXCR4 (Fig. 3). This indicated that gCXCR4 might be a common 234
ancestor to other CXCR4 proteins. 235
3.3. in vitro and in vivo expression of gCXCR4 236
To clarify the role of gCXCR4 in grouper, the expression of gCXCR4 in different 237
growth stages of grouper was measured by real-time PCR. gCXCR4 can be detected in all 238
examined fish samples (from 1dph to 40 dph). The expression levels of gCXCR4 was up 239
regulated and fluctuated in the period 1-4 dph and 6-8 dph , the gCXCR4 expressions are < 240
50. Two higher expression peaks (>100, p < 0.05) were observed at 18 dph and 38 dph (Fig. 241
4). 242
To evaluate the effect of gCXCR4 overexpression on cell proliferation, GF-1 cells 243
were transfected by pcDNA3.1-gCXCR4-GFP or pcDNA3.1-GFP overexpression vector. 244
The results showed that overexpression of gCXCR4-GFP for 12 h, 24 h, and 36 h had no 245
significant effects on cell viability (Fig. 5) but significantly repressed after 48 h (p < 0.05) 246
(Fig. 5) (n=5 per group). 247
3.4. Expression patterns of gCXCR4 on head kidney and intestine 248
gCXCR4 was mainly expressed in the head kidney (Fig. 6A) and on the surface of intestinal 249
villi of intestine (Fig. 6B). 250
3.5 The expressions of gCXCR4 in different organs of grouper 251
gCXCR4 was highly expressed in eye, gill, brain and important immune organs such as 252
15
spleen and head kidney (Fig. 7). Higher levels of expression were detected in eye, gill, 253
spleen, brain and head kidney tissues. Lower levels of expression were detected in fin, 254
muscle and heart tissues. Barely any gCXCR4 transcript was detected in the intestine (Fig. 255
7B). 256
3.6 The expressions of gCXCR4 after LPS or NNV challenge 257
The expression level of gCXCR4 in spleen was significantly increased after 6 h 258
post-injection of LPS (p < 0.05) and decreased at 24 h and 48 h post-injection of LPS (p < 259
0.05). At 72 h post-injection, gCXCR4 had returned to the base level as the control (Fig. 260
8A). 261
Forty-eight h after NNV infection, the juvenile groupers exhibited abnormal behaviors 262
such as loss of equilibrium and spiral swimming pattern and NNV can be detected at 72 h. 263
gCXCR4 expression was also up-regulated at the time point which was 72 h post-NNV 264
infection in eyes (p < 0.05) (Fig. 8B). 265
16 4. Discussion
267
The chemokine system has an important role in the host immune response against microbial 268
pathogens and provides a link between innate and adaptive immunity (Murphy et al., 2000). 269
The similar structure of gCXCR4 to other species (Alabyev et al., 2000; Tsuchida et al., 270
2007; Jia and Zhang, 2009) contains seven transmembrane regions, four extracellular 271
regions and four intracellular regions, and a conserved DRY motif (Fig. 2). The predicted 272
function of the gCXCR4 DRY motif was supported by the results of amino acid sequence 273
alignments of gCXCR4 and CXCR4 of other species, which function had been 274
demonstrated as important to G protein coupling (Doranz et al., 1999). The transmembrane 275
regions as well as the cysteine residue positions in the extracellular regions appear to be 276
highly conserved in CXCR4 evolution (Federsppiel et al., 1993; Heesen et al., 1996; 277
Alabyev et al., 2000; Tsuchida et al., 2007; Jia and Zhang, 2009). The posttranslational 278
modification, i.e. the tyrosine residues of the N terminus are sulfated in Golgi, of human 279
CXCR4 plays a crucial role on the infective ability of HIV (Farzan et al., 2002). However, 280
these tyrosine residues were not conserved in gCXCR4 (Figs.1 and 2), suggesting that the 281
posttranslational modification of gCXCR4 N-terminus is different. In addition, many serine 282
and threonine residues were identified in the C-terminus of gCXCR4 and might have the 283
modifications, i.e. phosphorylated as a prerequisite of signal transfer (Berson et al., 1996), 284
like other protein in CXCR4 family. 285
17
CXCR4 is expressed mainly in immune organs and central nervous system: thymus 286
and spleen of mouse (Heesen et al., 1996), chicken bursa (Liang et al., 2001), primate 287
(Macaca mulatta) brain (Federsppiel et al., 1993) and cattle locus coeruleus, cerebellum and 288
pons (Rimland et al., 1991). In grouper, gCXCR4 was highly expressed in NNV major target 289
organs, such as eyes and brain, and major lymphoid organs, such as gill, spleen and head 290
kidney. This also been reported in other fish species (Daniels et al., 1999; Jia and Zhang, 291
2009) in which CXCR4 expressed in central nervous system and immune system. 292
Interestingly, the expression of gCXCR4 in eye other than the immune related organs or 293
central nervous system has never been reported which raised the other possible function of 294
gCXCR4. Accordingly the grouper major lymphoid organs such as spleen, head kidney, gill 295
and mucosa-associated tissues appeared to be regions of gCXCR4 overproduction (Press 296
and Evensen., 1999). Furthermore, CXC chemokine system originates from the central 297
nervous system and may participate in central nervous system development (Huising et al., 298
2003). 299
SDF1/CXCR4 signaling plays a critical role in embryonic development and is essential 300
for development of cardiovascular, central nervous system, bone marrow colonization and 301
hematopoiesis in mice (Ma et al., 1998; Tachibana et al., 1998). In fish, CXCR4 has been 302
found in the early stage of zebrafish embryo and related to tissue polarity (Chong et al., 303
2001; Haas and Gilmour, 2006). The gene expression of gCXCR4 was highly expressed in 304
18
the period day14-day20 and day34-day40 larva that is coincided with dorsal spine formation 305
and pigmentation (Katsutoshi and Hiroshi, 2009). The results implied that chemokine 306
system exist in early developmental stage and play a key role in grouper development. 307
Immunohistofluorescence staining suggested that the protein gCXCR4 is expressed in 308
lymphoid organs (Fig. 6A) and mainly on the surface of intestinal villi. This may be due to 309
eyes, gills and surface of intestinal villi are continuously exposed to an environment which 310
may have potentially pathogenic microbes. LPS is an endotoxin constituent of the outer 311
membrane of Gram-negative bacteria which can induce immune responses and 312
inflammation (Raetz et al., 2008). In fish, it has been demonstrated that LPS can stimulate 313
the proliferation of neutrophils, monocytes, B lymphocytes and macrophages in a response 314
against LPS-induced inflammation (Swain et al., 2008). The results shown that LPS can 315
up-regulate the expression of gCXCR4 in the spleen and this also been showed in head 316
kidney and spleen of turbot after challenging with Vibrio harveyi (Jia et al., 2009). 317
gCXCR4 mRNA was up-regulated at 3 days post-infection and was significantly 318
increased in the eyes (Fig. 8B), suggesting that gCXCR4 is not only involved in the 319
response to bacteria invasion, but also have response to NNV infection. Although the eye 320
has been known to express chemokine receptors, such as CXCR1 and CXCR2 in mammals 321
(Goczalik et al., 2008), our detection of abundant gCXCR4 in the organs and significantly 322
up-regulated in NNV-infected fish was unexpected. Interestingly, grouper eye is one of the 323
19
main organs for NNV replication (Munday et al., 2002) and the immune response of NNV 324
infection involving macrophage-like cells and lymphocytes migrate to the eyes (Grotmol et 325
al., 1997; Nilsen et al., 2001; Munday et al., 2002). Taking those results together, we 326
hypothesized that the gCXCR4 expression is related to NNV infection and which may cause 327
by the immune related cells migration. However, too much gCXCR4 in cell could result in 328
significant growth obstruction which might due to the other functions of CXCR4 (Bleul et 329
al., 1996; Ganju et al., 1998). 330
In summary, our data indicated that the expression of grouper CXCR4 is regulated by 331
LPS or NNV challenge, and is expressed during embryogenesis, speculating its importance 332
in both immune and early developmental stage. The characterization of gCXCR4 between a 333
teleost fish and mammalians has provided valuable information for future functional 334
analysis of the gene. 335
20 Acknowledgements
337
We thank Dr. Brian D. Hoyle for editing the manuscript. This research was supported 338
by the National Science Council (NSC97-2313-B-006-001-MY3), and the Landmark 339
Project (B0127) of National Cheng Kung University, the plan of University Advancement, 340
Ministry of Education, Taiwan. 341
21 References
342
Alabyev, B.Y., Najakshin, A.M., Mechetina, L.V., Taranin, A.V., 2000. Cloning of a CXCR4 343
homolog in chondrostean fish and characterization of the CXCR4-specific structural 344
features. Dev Comp Immunol 24, 765-770. 345
Baldwin, J.M., 1994. Structure and function of receptors coupled to G proteins. Curr Opin 346
Cell Biol 6, 180-190. 347
Berson, J.F., Long, D., Doranz, B.J., Rucker, J., Jirik, F.R., Doms, R.W., 1996. A 348
seven-transmembrane domain receptor involved in fusion and entry of T- cell-tropic 349
human immunodeficiency virus type 1 strains. J Virol 70, 6288-6295. 350
Bleul, C.C., Farzan, M., Choe, H., Parolin, C., Clark-Lewis, I., Sodroski, J., Springer, T. A., 351
1996. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks 352
HIV-1 entry. Nature 6594, 829-833. 353
Chen, Y.M., Su, Y.L., Shie, P.S., Hwang, S.L., H.L., Chen, T.Y., 2008. Grouper Mx confers 354
resistance to nodavirus and interacts with coat protein. Dev Comp Immunol 32, 355
825-836. 356
Chen, Y.M., Kuo, C.E., Wang, T.Y., Shie, P.S., Wang, W.C., Huang, S.L., Tsai, T.J., Chen, 357
P.P., Chen, J.C., Chen, T.Y., 2010. Cloning of an orange-spotted grouper Epinephelus 358
coioides heat shock protein 90AB (HSP90AB) and characterization of its expression 359
in response to nodavirus. Fish Shellfish Immunol 28, 895-904. 360
22
Chong, S.W., Emelyanov, A., Gong, Z., Korzh, V., 2001. Expression pattern of two 361
zebrafish genes, cxcr4a and cxcr4b. Mech Dev 109, 347-354. 362
Daniels, G.D., Zou, J., Charlemagne, J., Partula, S., Cunningham, C., Secombes, C.J., 1999. 363
Cloning of two chemokine receptor homologs (CXC-R4 and CC-R7) in rainbow trout 364
Oncorhynchus mykiss. J Leukoc Biol 65, 684-690. 365
Doranz, B.J., Orsini, M.J., Turner, J.D., Hoffman, T.L., Berson, J.F., Hoxie, J.A., Peiper, 366
S.C., Brass, L.F., Doms, R.W., 1999. Identification of CXCR4 domains that support 367
coreceptor and chemokine receptor functions. J Virol 73, 2752-2761. 368
Farzan, M., Babcock, G.J., Vasilieva, N., Wright, P.L., Kiprilov, E., Mirzabekov, T., Choe, 369
H., 2002. The role of post-translational modifications of the CXCR4 amino terminus 370
in stromal-derived factor 1 alpha association and HIV-1 entry. J Biol Chem 277, 371
29484-29489. 372
Federsppiel, B., Melhado, I.G., Duncan, A.M.V., Delaney, A., Schappert, K., Clark-Lewis, I., 373
Jirik, F.R., 1993. Molecular cloning of the cDNA and chromosomal localization of the 374
gene for a putative seven-transmembrane segment (7-TMS) receptor isolated from 375
human spleen. Genomics 16, 707-712. 376
Feng, Y., Broder, C.C., Kennedy, P.E., Berger, E.A., 1996. HIV-1 entry cofactor: functional 377
cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272, 378
872-877. 379
23
Ganju, R.K., Brubaker, S.A., Meyer, J., Dutt, P., Yang, Y., Qin, S., Newman, W., Groopman, 380
J.E., 1998. The alpha-chemokine, stromal cell-derived factor-1alpha, binds to the 381
transmembrane G-protein-coupled CXCR-4 receptor and activates multiple signal 382
transduction pathways. J Biol Chem 273, 23169-23175 383
Goczalik, I., Ulbricht, E., Hollborn, M., Raap, M., Uhlmann, S., Weick, M., Pannicke, T., 384
Wiedemann, P., Bringmann, A., Reichenbach, A., Francke, M., 2008. Expression of 385
CXCL8, CXCR1, and CXCR2 in neurons and glial cells of the human and rabbit 386
retina. Invest Ophthalmol Vis Sci 49, 4578–4789. 387
Grotmol, S., Totland, G.K., Torud, K., Hjeltnes, B.K., 1997. Vacuolating encephalopathy 388
and retinopathy associated with a nodavirus-like agent: a probable cause of mass 389
mortality of cultured larval and juvenile Atlantic halibut Hippoglossus hippoglossus. 390
Dis Aquat Organ 29, 85-97. 391
Haas, P., Gilmour, D., 2006. Chemokine signaling mediates self-organizing tissue migration 392
in the zebrafish lateral line. Dev Cell 10, 673-680. 393
Heesen, M., Berman, M.A., Benson, J.D., Gerard, C., Dorf, M.E., 1996. Cloning of the 394
mouse fusin gene, homologue to a human HIV-1 co-factor. J Immunol 157, 395
5455-5460. 396
Hesselgesser, J., Taub, D., Baskar, P., Greenberg, M., Hoxie, J., Kolson, D. L., Horuk, R., 397
1998. Neuronal apoptosis induced by HIV-1 gp120 and the chemokine SDF-1 alpha is 398
24
mediated by the chemokine receptor CXCR4. Curr Biol 10, 595-598. 399
Horuk, R., 2009. Chemokine receptor antagonists: overcoming developmental hurdles. Nat 400
Rev Drug Discov 8, 23-33. 401
Huising, M.O., Stet, R.J., Kruiswijk, C.P., Savelkoul, H.F., Lidy Verburg-van Kemenade, 402
B.M., 2003. Molecular evolution of CXC chemokines: extant CXC chemokines 403
originate from the CNS. Trends Immunol 24, 307-313. 404
Jia, A., Zhang, X.H., 2009. Molecular cloning, characterization, and cxpression analysis of 405
the CXCR4 gene from turbot: Scophthalmus maximus. J Biomed Biotechnol 406
doi:10.1155/2009/767893. 407
Katsutoshi, K., Kohno, H., 2009. Morphological development of larval and juvenile 408
blacktip grouper, Epinephelus fasciatus. Fish Sci 75, 1239–1251. 409
Kim, C.H., Broxmeyer, H.E., 1999. Chemokines: signal lamps for trafficking of T and B 410
cells for development and effector function. J Leukoc Biol 65, 6-15. 411
Kuo, H.C., Wamg, T.Y., Chen, P.P., Chen, Y.M., Chuang, H.C., Chen T.Y., 2011. Real-time 412
quantitative PCR asaay for monitoring of nervous necrosis virus infection in grouper 413
aquaculture. J Clin Microbiol 49, 1090-1096. 414
Liang, T.S., Hartt, J.K., Lu, S., Martins-Green, M., Gao, J.L., Murphy, P.M., 2001. Cloning, 415
mRNA distribution, and functional expression of an avian counterpart of the 416
chemokine receptor/HIV coreceptor CXCR4. J Leukoc Biol 69, 297-305. 417
25
Ma, Q., Jones, D., Borghesani, P.R., Segal, R. A., Nagasawa, T., Kishimoto, T., Bronson, 418
R.T., Springer, T.A., 1998. Impaired B-lymphopoiesis, myelopoiesis, and derailed 419
cerebellar neuron migration in CXCR4- and SDF-1-deficient mice. Proc Natl Acad 420
Sci U S A 95, 9448-9453. 421
Munday, B.L., Kwang, J., Moody, N., 2002. Betanodavirus infections of teleost fish: a 422
review. J Fish Dis 25, 127-142. 423
Murphy, P.M., Baggiolini, M., Charo, I.F., Hebert, C.A., Horuk, R., Matsushima, K., Miller, 424
L.H., Oppenheim, J.J., Power, C.A., 2000. International union of pharmacology. XXII. 425
nomenclature for chemokine receptors. Pharmacol Rev 52, 145-176. 426
Nilsen, R., Ranheim, T., Hansen, M.K., Taksdal, T., Totland, G.K., 2001. Pathology in 427
persistent nodavirus infected juvenile Atlantic halibut Hippoglossus hippoglossus. In: 428
Tenth International Conference of the European Association of Fish Pathologists 429
Abstract P-117, 9-14. 430
Olson, T.S., Ley, K., 2002. Chemokines and chemokine receptors in leukocyte trafficking. 431
Am J Physiol Regul Integr Comp Physiol 283, R7-28. 432
Press, C. M.,and Evensen, Ø., 1999. The morphology of the immune system in teleost fishes. 433
Fish Shellfish Immunol 9, 309-318. 434
Raetz, C.R., Whitfield, C., 2002. Lipopolysaccharide endotoxins. Annu Rev Biochem. 71, 435
635-700. 436
26
Rimland, J., Xin, W., Sweetnam, P., Saijoh, K., Nestler, E.J., Duman, R.S., 1991. Sequence 437
and expression of a neuropeptide Y receptor cDNA. Mol Pharmacol 40, 869-875. 438
Swain, P., Nayak, S.K., Nanda, P.K., Dash, S., 2008. Biological effects of bacterial 439
lipopolysaccharide (endotoxin) in fish: a review. Fish Shellfish Immunol 25, 191-201. 440
Tachibana, K., Hirota, S., Iizasa, H., Yoshida, H., Kawabata, K., Kataoka, Y., Kitamura, Y., 441
Matsushima, K., Yoshida, N., Nishikawa, S., Kishimoto, T., Nagasawa, T., 1998. The 442
chemokine receptor CXCR4 is essential for vascularization of the gastrointestinal 443
tract. Nature 393, 591-594. 444
Tsuchida, S., Kagi, A., Takahashi, T., 2007. Characterization of cDNA and genomic 445
sequences encoding a canine chemokine receptor, CXCR4 and its ligand CXCL12. 446
Vet Immunol Immunopathol 116, 219-225. 447
Valentin, G., Haas, P., Gilmour, D., 2007. The chemokine SDF1a coordinates tissue 448
migration through the spatially restricted activation of Cxcr7 and Cxcr4b. Curr Biol 449
17, 1026-1031. 450
27 Legends of figures
452
Fig. 1. Nucleotide sequence and deduced amino acid sequence of the open reading 453
frame of Epinephelus coioides cDNA. The bold letters represent the start codon (ATG) and 454
the stop codon (TAG). The regions of seven-transmembrane domains are at amino acids 455
50 –72, 85–104, 119–141, 161–180, 216–238, 259–281 and 301–323. The Genbank 456
accession number of gCXCR4 is HQ185191. 457
Fig. 2. Protein alignment and analysis of gCXCR4 with homologues from other species. 458
The other species used for comparison were Homo sapiens (GenBank accession number 459
CAA12166), Mus muluscus (GenBank accession number AAH31665), Cyprinus carpio, 460
Oncorhynchus mykiss (GenBank accession number CAA04493) and Psetta maxima 461
(GenBank accession number ABP48751). The black letters denote the consensus sequence 462
of CXCR4 of the different species. The bars represent transmembrane region of gCXCR4 463
determined using the TMHMMM program in the ExPASy Proteomics Server database. The 464
DRY motif is boxed. The asterisk represents conserved cysteine residue on each of the four 465
extracellular domains that located at positions 33aa, 118aa, 198aa and 292aa. 466
Fig. 3. Phylogenetic analysis of CXCR4 protein family members. The amino acids of the 467
different CXCR4 species obtained from the NCBI GenBank were aligned using ClustalW. 468
The Neighbor-Joining tree was created by MEGA4.0 software with a bootstrap value of 469
1,000. Accession numbers of chemokine receptors amino acid sequences obtained from 470
28
GenBank were: Homo sapiens CXCR1 NP_000625; Mus musculus CXCR1 NP_839972; 471
Cyprinus carpio CXCR1 BAA31458; Takifugu rubripes CXCR1 NP_001072110; Homo 472
sapiens CXCR2 NP_001161770; Mus musculus CXCR2 NP_034039; Bos taurus CXCR2 473
ABC59060; Homo sapiens CXCR3 EAX05283; Danio rerio CXCR3a NP_001082899; 474
Ctenopharyngodon idella CXCR3 AAW69766; Petromyzon marinus CXCR4 AAO21209; 475
Epinephelus coioides CXCR4 HQ185191; Homo sapiens CXCR4 CAA12166; Mus 476
musculus CXCR4 AAH98322; Bos taurus CXCR4 NP_776726; Sus scrofa CXCR4 477
AAZ32767; Cyprinus carpio CXCR4 BAA32797; Oncorhynchus mykiss CXCR4 478
CAA04493; Salmo salar CXCR4 BT060355; Danio rerio CXCR4 AAF1756; Ictalurus 479
punctatus CXCR4 ACS45337; Acipenser ruthenus CXCR4 CAB60252; Xenopus laevis 480
CXCR4 AAI10722; Oryzias latipes CXCR4 ABC41565; Psetta maxima CXCR4 481
ABP48751; Homo sapiens CXCR5 AAI10353; Mus musculus CXCR5 AAH64059; 482
Ctenopharyngodon idella CXCR5 ACZ06880; Mus musculus CXCR6 NP_109637; Homo 483
sapiens CXCR6 NP_006555; Bos taurus CXCR6 NP_001014859; Homo sapiens CXCR7 484
NP_064707; Mus musculus CXCR7 NP_031748 and Xenopus laevis CXCR7 485
NP_001082236. 486
Fig. 4. Gene expression profile of gCXCR4 was examined in different development 487
stages of Epinephelus coioides. The total RNA was isolated from different stages and gene 488
expression of gCXCR4 was determined by real-time PCR. 489
29
Fig. 5. Effects of overexpression of gCXCR4 on GF-1 cell proliferation. The cells 490
proliferation was quantified by measuring MTT absorbance at 570 nm. Vertical bars 491
indicate the mean ± S.E (N=3). **p < 0.01. The Blank was the spontaneous proliferation of 492
GF-1 cells without treating any plasmid; the GFP was the group transfected with the same 493
backbone of the plasmid to the gCXCR4-GFP group without inserting the gCXCR4. 494
Fig. 6. Expression of gCXCR4 in (A) head kidney and (B) intestine of healthy grouper 495
using immunohistofluorescence staining. (a) and (d): Nucleus was detected using Hoechst 496
33342 (blue). (b) Control experiments were carried out with control rabbit antiserum as the 497
primary antiserum, and visualized with Alexa Fluor® 594 goat anti-rabbit IgG (H+L) (red). 498
(e) The expression of gCXCR4 was detected using rabbit anti-gCXCR4 antiserum and 499
visualized with Alexa Fluor® 594 goat anti-rabbit IgG (H+L). (c) Merged image from 500
figures (a) and (b). (f) Merged image from figures (d) and (e). Bars = 1 mm (A) and 50 μm 501
(B). 502
Fig. 7. Gene expression of gCXCR4 in different tissues. (A) RT-PCR and (B) real-time 503
PCR analysis of gCXCR4 gene expression in different tissues including eye, fin, gill, muscle, 504
head kidney, heart, spleen ,intestine and brain of healthy adult grouper. β-actin 505
amplification was used as an internal control. Vertical bars indicate the mean ± S.E (N=3). 506
*p < 0.05. 507
Fig. 8. Expression level of gCXCR4 mRNA in grouper after challenge with LPS (A) 508
30
and NNV (B). (A) Relative expression level of gCXCR4 mRNA in spleen of grouper after 509
challenge with LPS or PBS. (B) Analysis of expression of gCXCR4 gene in control or NNV 510
infected juvenile grouper or eye of juvenile grouper groups by real-time RT-PCR. gCXCR4 511
mRNA levels (relative to β-actin mRNA) between different time were compared by the 512
t-test. Vertical bars indicate the mean ± S.E (N=3). *p < 0.05. 513
31
Figure 1
32
Figure 2
33
Figure 3
34
Figure 4
35
Figure 5
36
Figure 6
37
Figure 7
38
Figure 8
39
Table1. Primers used in this study
Name Sequence gCXCR4-5SP1 CAGTAACACACAAGGAGGACCAG gCXCR4-5SP2 GCTGATGAATGCCAGGATGAGCACACT gCXCR4-5SP3 AGCAGCAGAGAGATGTAGCCG gCXCR4-3SP1 CGCTACCTGGCTGTAGTCCGA gCXCR4-3SP2 CTTCGTGTGCTGGCTGCCGTATGG gCXCR-F ATGTCGTACTATGAGCATATCGTCTTCG gCXCR-R CTAGCTGGAATGTAAACTGGAGGACTC gCXCR4-GFP-R GGCTGGAATGTAAACTGGAGGACTC gCXCR4-EXI-F ATCGTAGGATCCGATGTCGTACTATGAGCAT gCXCR4-EXI-R ACGATGTCGACAGGTAAGAAGACCTGCTGGAG gCXCR4-EXIII-F ATCGTAGTCGACCCGGACTTGATCTATGCCC gCXCR4-EXIII-R ACGATCTCGAGGTGGAAGACTGCAACCCAG Q-gCXCR4-F CACACTGCTGCCTGAACCCACTGCT Q-gCXCR4-R CTAGCTGGAATGTAAACTGGAGGACTC β-actin-F TGCCTCTGGTCGTACCACTGGTATTGTC β-actin-R GGCAGCAGTGCCCATCTCCTGCTCGA
40
Table 2. Protein sequences similarity* of CXCR4 from different species E. coioides H. sapiens M. musculus C. carpio D. rerio O. mykiss P. maxima Extracellular domains E. coioides 100 22.1 8.5 28.8 32.2 33.6 29.3 H. sapiens 100 15.9 30.3 36.3 31.9 30.3 M. musculus 100 13.7 16.2 14.6 14.9 C. carpio 100 75.5 43.4 33.1 D. rerio 100 51.4 37.3 O. mykiss 100 46.6 P. maxima 100 Transmembrane domains E. coioides 100 57.7 32.9 59.0 59.3 59.1 62.3 H. sapiens 100 35.4 71.6 70.7 75.0 75.2 M. musculus 100 37.9 36.0 37.3 37.9 C. carpio 100 92.3 80.1 84.0 D. rerio 100 77.8 82.7 O. mykiss 100 84.8 P. maxima 100 Intracellular domains E. coioides 100 43.6 15.8 44.0 45.4 41.4 38.8 H. sapiens 100 24.8 64.4 62.7 58.4 59.4 M. musculus 100 20.2 19.8 22.2 21.4 C. carpio 100 89.8 73.5 74 D. rerio 100 71.4 73.5 O. mykiss 100 76 P. maxima 100