JOURNAL OFBACTERIOLOGY,
0021-9193/98/$04.0010 Mar. 1998, p. 1338–1341 Vol. 180, No. 5
Copyright © 1998, American Society for Microbiology
Molecular Cloning and Characterization of Fengycin Synthetase
Gene fenB from Bacillus subtilis
GUANG-HUEY LIN,1,2CHYI-LIANG CHEN,2JOHANNES SCHENG-MING TSCHEN,3SAN-SAN TSAY,1 YU-SUN CHANG,2ANDSHIH-TUNG LIU2*
Graduate Institute of Botany, National Taiwan University, Taipei, 106,1Graduate Institute of Botany, National Chung-Hsing University, Taichung, 402,3and Molecular Genetics Laboratory,
Department of Microbiology and Immunology, Chang-Gung University, Kwei-Shan, Taoyuan, 333,2Taiwan
Received 3 November 1997/Accepted 30 December 1997
A fengycin synthetase gene, fenB, has been cloned and sequenced. The protein (FenB) encoded by this gene has a predicted molecular mass of 143.6 kDa. This protein was overexpressed in Escherichia coli and was purified to near homogeneity by affinity chromatography. Experimental results indicated that the recombinant FenB has a substrate specificity toward isoleucine with an optimum temperature of 25°C, an optimum pH of 4.5, a Kmvalue of 922mM, and a turnover number of 236 s21. FenB also consists of a thioesterase domain, suggesting that this protein may be involved in the activation of the last amino acid of fengycin.
Fengycin is a lipopeptidic antifungal antibiotic produced by
Bacillus subtilis F29-3 (2, 4), consisting of 10 amino acids and
having a primary sequence similar to that of plipastatin (10, 16, 24). Mutagenesis and sequencing studies found that fengycin is probably synthesized nonribosomally by peptide synthetases (1, 2). A peptide synthetase may consist of one to several amino acid activation modules for the activation of specific amino acids (9). In each module, there is an amino acid ad-enylation domain of approximately 500 amino acids, consisting of five highly conserved motifs for ATP binding and for ATPase activity (19). Mutation in the motifs can significantly reduce the activity of amino acid activation (6, 7), indicating that these motifs are indeed essential for peptide synthesis (7). In a peptide synthetase module, the C-terminal boundary of the activation domain is followed by a thioester formation domain which contains a conserved DNFYxLGGHSL motif for the binding of cofactor 49-phosphopantetheine (9, 19). Af-ter adenylation, the amino acid is transferred to the 49-phos-phopantetheine at the carrier domain (20). A transpeptidation step subsequently follows, which transfers the amino acid on the cofactor of the initiating module to the activated amino acid at the thioester formation domain in the next module to form a peptide (9). This condensation step continues from one module to the other until a complete peptide is synthesized (9). It is thought that peptide synthetases may form a complex in vivo and the amino acid activation modules among the en-zymes are connected and aligned colinearly with the sequence of the amino acids in the antibiotic (8, 18), thereby allowing an antibiotic with the correct sequence to be sequentially synthe-sized. A peptide synthetase also consists of a conserved spacer domain which is present at the N-terminal region, upstream from the adenylation domain of each module (4), except for the module activating the initiating amino acid in which the spacer domain is located in the C-terminal end, downstream from the thioester carrier domain (20). In addition, the C terminus of the last module of a peptide synthetase may con-tain an epimerization domain for the conversion ofL-amino
acid to D-amino acid (4) and a spacer domain which may be
essential for the elongation of peptide. The peptide synthe-tases involved in the activation of the last amino acid of a peptide usually consist of a thioesterase-like domain in the C-terminal region (3). This domain may be responsible for the release of the peptide from 49-phosphopantetheine, a prereq-uisite for terminating nonribosomal peptide synthesis (18). In this study, we have cloned, sequenced, and characterized a fengycin synthetase gene, fenB. This gene is involved in the activation of the last amino acid of fengycin.
Nucleotide sequence of fenB. In a previous study (2), we identified a 46-kb cosmid clone, pFC660, which contains genes encoding fengycin synthesis. This cosmid consists of three
BamHI fragments—B1 (18 kb), B2 (12 kb), and B3 (16 kb) (2).
In this study, we have sequenced the entire B2 fragment and found that this fragment is actually 11,459 bp long. In the 39 portion of the fragment, there is a 3,825-bp gene, fenB, which is preceded by a ribosomal binding site and is followed by a putative transcriptional stop signal, which consists of a stem-loop structure and a stretch of T’s. The 59 portion of the B2 fragment, ranging from nucleotides (nt) 1 to 6,036, consists of an incomplete open reading frame, which is actually the 39 portion of a 10,488-bp peptide synthetase gene, fenA. The
* Corresponding author. Molecular Genetics Laboratory, Depart-ment of Microbiology and Immunology, Chang-Gung University, Kwei-Shan, Taoyuan, 333, Taiwan. Phone: 3-328-0292. Fax: 886-3-328-0292. E-mail: cgliu@cguaplo.cgu.edu.tw.
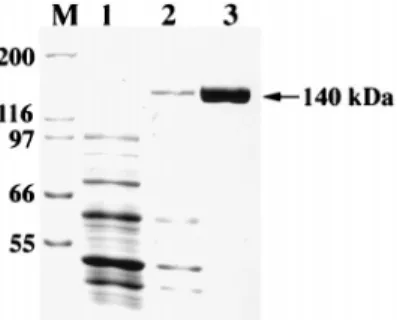
FIG. 1. Expression and purification of His-tagged recombinant FenB. Cell extracts obtained from cells before (lane 1) and after (lane 2) IPTG induction and proteins eluted from His-Bind column (lane 3) were analyzed by SDS-PAGE and stained by Coomassie blue. The top band in lane 2 is overexpressed FenB (140 kDa). The positions of molecular mass markers (M) (in kilodaltons) are shown to the left of the gel.
1338
at NATIONAL TAIWAN UNIV MED LIB on May 3, 2009
jb.asm.org
protein encoded by fenB (FenB) consists of six core sequences (Table 1) and a thioesterase-like domain (GYSAG) which are highly conserved among peptide synthetases (3, 5). The fenB sequence shows 80.6% homology to a gene in the pps operon of B. subtilis 168 (21). Since B. subtilis 168 does not produce fengycin, it is unclear whether the fenB-like gene in strain 168 is functional or whether the proteins encoded by these two genes have the same function.
Expression and purification of FenB.To obtain a sufficient amount of FenB for enzyme analysis, we overexpressed fenB in
Escherichia coli M15(pRep4) (Qiagen, Hilden, Germany). This
overexpression was accomplished by cloning fenB into an ex-pression vector, pQE60 (Qiagen). The fenB DNA (nt 1 to 3822) was amplified by using primers B1 (59-ATCCATGGTT
AAAAACCAAAAAAAT) and B2 (59-ACGGATCCATGCT TATTTGGCAGC), which contained an NcoI restriction site and a BamHI restriction site at 59 ends, respectively. PCR was then performed for 30 cycles, with 1 cycle consisting of 1 min at 94°C, 2 min at 40°C, and 3 min at 72°C. The amplified fragment was cut by BamHI and NcoI and was inserted into the
NcoI-BamHI sites of pQE60. FenB expression was induced by
isopropyl-b-D-thiogalactopyranoside (IPTG) treatment. For
the purification of FenB, cells were frozen in liquid nitrogen and then were thawed at room temperature. A total of three cycles of freeze-thawing were conducted. Cells were suspended in 4 ml of buffer containing 5 mM imidazole, 0.5 mM NaCl, and 20 mM Tris-HCl (pH 7.9) and were sonicated at 0°C for 48 5-s pulses at 10-s intervals with an output control setting at 3 with a sonicator (model UP400A; Ultrasonic Processor Corp., Copiague, N.Y.). Next, cell extract was centrifuged at 15,000 rpm for 60 min at 4°C with a Sorvall SS-34 rotor. FenB in the supernatant was then purified with a His-Bind column (Nova-gen, Madison, Wis.) (1.5 by 4 cm), and FenB in the fractions was examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12) and by staining with Coo-massie blue (Merck, Darmstadt, Germany). The expressed FenB has a molecular mass of 140 kDa, as determined by SDS-PAGE (Fig. 1, lanes 2 and 3). The chromatography pro-cedure was able to purify FenB to near homogeneity (Fig. 1, lane 3). In addition, approximately 300 mg of recombinant FenB could be purified from 50 ml of culture.
Substrate specificity.The enzymatic activity of recombinant FenB was determined by an ATP-PPi exchange assay (14) using a reaction mixture containing [32P]tetrasodium
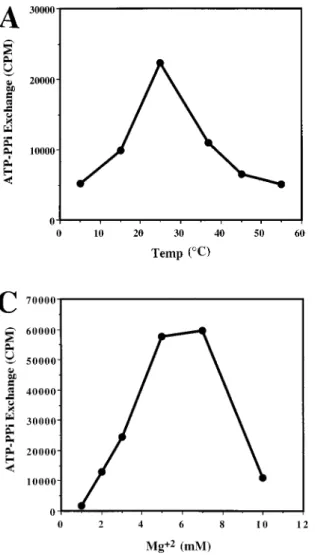
pyro-FIG. 2. Properties of FenB. Effects of temperature (A), pH (B), and Mg21 concentration (C) in FenB activity. The optimum Mg21concentration was de-termined in a buffer containing 2 mM EDTA.
TABLE 1. Comparison of the amino acid sequences conserved in peptide synthetases and FenB
Motifa Conserved sequencea Functiona Sequence in FenB Positions in fenB (nt)
Spacer HHILxDGW Unknown HHILMDGGW 435–459
Core 1 LKAGGAYVPID Unknown LKAGGTYLPLD 1,617–1,650
Core 2 YSGTTGxPKGV ATP binding SSGSTGRPKGV 1,851–1,881
Core 3 GELCIGGxGxARGYL ATP binding GELCVGGEGVAKGYL 2,409–2,454
Core 4 YxTGD ATPase YRTGD 2,517–2,532
Core 5 VKIRGxRIELGEIE ATP binding IKIRGKRIEPAEIE 2,559–2,631
Core 6 DNFYxLGGHSL 49-Phosphopantetheine binding
(thioester formation) DFFALGGHSL 2,982–3,012
aData taken from Stachelhaus and Marahiel (18, 19).
VOL. 180, 1998 NOTES 1339
at NATIONAL TAIWAN UNIV MED LIB on May 3, 2009
jb.asm.org
phosphate (9,120 Ci/mmol) (NEN, Boston, Mass.), 2 mM ATP, and 2 mM amino acid. The amino acids used for the assay included the eight amino acids present in the fengycin molecule (Table 2) (10, 25) as well as other common amino acids. Experimental results indicated that adding isoleucine to the reaction mixture produced the highest ATP-PPiexchange activity (Table 2), i.e., approximately 8- to 70-fold higher than the binding to the other amino acids. Above results suggested that FenB has a substrate specificity toward isoleucine. Previ-ous reports have demonstrated that isoleucine is the last amino acid of the fengycin molecule (10, 25). This finding suggests that FenB is not only responsible for activating the last amino acid of fengycin but also involved in releasing the fengycin molecule from the peptide synthetase. Actually, the amino acid sequence of FenB also reveals that this is indeed the case. In the C-terminal region of FenB, the protein consists of a thio-esterase-like domain instead of an epimerase domain, a fea-ture shared by all the peptide synthetases involved in activating the last amino acid of antibiotics (3, 5).
Binding of amino acid to FenB.Covalent binding of amino acid to FenB was examined with 1 mCi of L-[14C]isoleucine
(315 mCi/mmol) (Amersham, Buckinghamshire, England) and 11mg of purified recombinant FenB by the method described by Ullrich et al. (23). The reaction was allowed to proceed for 30 min at 37°C and then was stopped by adding 2 ml of ice-cold 10% trichloroacetic acid. Our results demonstrated that [14 C]-isoleucine could bind to FenB covalently and gave a radioac-tivity reading of 6,725 cpm, whereas the negative control, which lacked ATP in the reaction mixture, had a value of 98 cpm. This binding is specific, since nonradioactive isoleucine, when added in an excessive amount in the reaction mixture, could compete with the binding of radioactive isoleucine to FenB. In theory, binding of isoleucine to FenB requires a prior binding of 49-phosphopantetheine to the enzyme (11, 17). In the case of surfactin synthetases, this binding is catalyzed by the enzyme encoded by the sfp gene in B. subtilis (13). A similar gene is also involved in plipastatin synthesis (22). A previous study has demonstrated that approximately 14% of the peptide synthetase expressed in E. coli has a phosphopantetheinyl group attached to the enzyme (19). This binding is catalyzed by an E. coli enzyme, phosphopantetheinyl transferase (17, 19). Presumably, the phosphopantetheinyl group of coenzyme A is transferred to FenB by the same mechanism and subsequently results in the binding of the [14C]isoleucine to the enzyme. We found that approximately 13% of FenB expressed in E. coli bound to the amino acid.
Biochemical characterization of recombinant FenB.The re-combinant FenB enzyme had optimum activity at 25°C (Fig. 2A), at pH 4.5 (Fig. 2B), and with a Mg21 concentration between 5 and 8 mM in a buffer containing 2 mM EDTA (Fig.
2C). The activity of FenB at pH 7.0 is approximately 18-fold lower than the activity exhibited under pH 4.5 (Fig. 2B). The low optimum pH for FenB may allow the enzyme to function efficiently in the acidic intracellular environment. Although many peptide synthetases have been isolated from Bacillus spp. and characterized biochemically (11, 15, 19), the optimum pHs of these enzymes were not determined in those studies. The activity also decreased when the Mg21concentration exceeded 10 mM (Fig. 2C). It is likely that a high concentration of Mg21 affects the amount of EDTA, which may be critical in main-taining the enzyme’s stability. The recombinant FenB, under optimum conditions, exhibited Michaelis-Menten kinetics, with a Kmfor isoleucine of 922mM and a turnover number of
236 s21.
In summary, we have sequenced and characterized the fengycin synthetase gene fenB from B. subtilis F29-3. Experi-mental results demonstrate that the FenB protein functions as a peptide synthetase which is involved in the nonribosomal synthesis of fengycin. This enzyme is responsible for the ad-enylation of isoleucine and for the binding of the amino acid to its cofactor, 49-phosphopantetheine. Evidence presented herein suggests that FenB is involved in the activation of the last amino acid of the fengycin peptide. Our results should provide a valuable reference for future studies involving fengy-cin synthesis.
Nucleotide sequence accession numbers.The nucleotide se-quences of the 3,825-bp fenB gene and the 10,488-bp fenA gene have been deposited in GenBank under accession no. L42523 and AF023464, respectively.
We thank Hans von Do¨hren and J.-S. Yu for their technical advice. This research was supported by Medical Research Grant CMRP525 from the Chang-Gung Memorial Hospital and by Biological Research Grant NSC-86-2314-B-182-028 from the National Science Council of the Republic of China.
REFERENCES
1. Chang, L. K., C. L. Chen, Y. S. Chang, J. S. M. Tschen, J. M. Chen, and S. T.
Liu.1994. Construction of Tn917ac1, a transposon useful for mutagenesis and cloning of Bacillus subtilis genes. Gene 150:129–134.
2. Chen, C. L., L. K. Chang, Y. S. Chang, S.-T. Liu, and J. S. M. Tschen. 1995. Transposon mutagenesis and cloning of the genes encoding the enzymes of fengycin biosynthesis in Bacillus subtilis. Mol. Gen. Genet. 243:121–125. 3. Cosmina, P., F. Rodriguez, F. de Ferra, G. Grandi, M. Perego, G. Venema,
and D. von Sinderen.1993. Sequence and analysis of the genetic locus responsible for surfactin synthesis in Bacillus subtilis. Mol. Microbiol. 8:821– 831.
4. de Cre´cy-Lagard, V., P. Marliere, and W. Saurin. 1995. Multienzymatic nonribosomal peptide biosynthesis: identification of the functional domains catalyzing peptide elongation and epimerization. Life Sci. 318:927–936. 5. de Ferra, F., F. Rodriguez, F. Tortora, C. Tosi, and G. Grandi. 1997.
Engi-neering of peptide synthetase. J. Biol. Chem. 272:25304–25309.
6. Elsner, A., H. Enger, W. Saenger, L. Hamoen, G. Venema, and F. Bernhard. 1997. Substrate specificity of hybrid modules from peptide synthetases. J. Biol. Chem. 272:4814–4819.
7. Gocht, M., and M. A. Marahiel. 1994. Analysis of core sequences in the D-phe activating domain of the multifunctional peptide synthetase TycA by site-directed mutagenesis. J. Bacteriol. 176:2654–2662.
8. Kleinkauf, H., and H. von Do¨hern. 1990. Nonribosomal biosynthesis of peptide antibiotics. Eur. J. Biochem. 192:1–15.
9. Kleinkauf, H., and H. von Do¨hren. 1996. A nonribosomal system of peptide biosynthesis. Eur. J. Biochem. 236:335–351.
10. Koch, U. 1988. Fengycin: Strukturaufkla¨rung eines mikroheterogene Li-popeptidolidantibiotikums. Dissertation, zur Erlangung des Grades eines Doktors, der Naturwissenschaften. Eberhard-Karls-Universita¨t zu Tu¨bingen, Tu¨bingen, Germany.
11. Ku, J., R. G. Mirmira, L. Liu, and D. L. Santi. 1997. Expression of a functional non-ribosomal peptide synthetase module in Escherichia coli by coexpression with a phosphopantetheinyl transferase. Chem. Biol. 4:203– 207.
12. Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685.
13. Lambalot, R. H., A. M. Gehring, R. S. Flugel, P. Zuber, M. LaCelle, M. A.
Marachiel, R. Reid, C. Khosla, and C. T. Walsh. 1996. A new enzyme
TABLE 2. ATP-PPiexchange activity of FenB
Amino acid Exchange activity (cpm)a % Activity
Isoleucine 107,038 100 Glutamic acid 284 0.26 Alanine 3,370 3.1 Proline 1,384 1.2 Ornithine 1,582 1.4 Threonine 2,171 2.0 Tyrosine 1,834 1.7 Valine 13,630 12.7 Blank 1,497 1.4
aThe reaction mixture consisted of 84mg of recombinant FenB. The reaction was allowed to proceed for 10 min.
1340 NOTES J. BACTERIOL.
at NATIONAL TAIWAN UNIV MED LIB on May 3, 2009
jb.asm.org
superfamily—the phosphopantetheinyl transferases. Chem. Biol. 3:923–936. 14. Lee, S. G., and F. Lipmann. 1975. Tyrocidine synthetase. Methods Enzymol.
43:585–602.
15. Mootz, H., and M. A. Maraheil. 1997. The tyrocidine biosynthesis operon of Bacillus brevis: complete nucleotide sequence and biochemical characteriza-tion of funccharacteriza-tional internal adenylacharacteriza-tion domains. J. Bacteriol. 179:6843–6850. 16. Nishikiori, T., H. Naganawa, Y. Muraoka, T. Aoyagi, and H. Umezawa. 1986. Plipastatins: new inhibitors of phospholipase A2, produced by Bacillus cereus BMG302-fF67. III. Structural elucidation of plipastatins. J. Antibiot. 39:755– 761.
17. Stachelhaus, T., A. Hu¨ser, and M. A. Marahiel. 1996. Biochemical charac-terization of peptidyl carrier protein (PCP), the thiolation domain of mul-tifunctional peptide synthetases. Chem. Biol. 3:913–921.
18. Stachelhaus, T., and M. A. Marahiel. 1995. Modular structure of genes encoding multifunctional peptide synthetase required for non-ribosomal peptide synthesis. FEMS Microbiol. Lett. 125:3–14.
19. Stachelhaus, T., and M. A. Marahiel. 1995. Modular structure of peptide synthetase revealed by dissection of the multifunctional enzyme GrsA. J. Biol. Chem. 270:6163–6169.
20. Stein, T., J. Vater, V. Kruft, A. Otto, B. Wittmann-Liebold, P. Franke, M.
Panico, R. McDowell, and H. R. Morris.1996. The multiple carrier model of nonribosomal peptide biosynthesis at modular multienzyme templates. J. Biol. Chem. 271:15428–15435.
21. Tognoni, A., E. Franchi, C. Magistrelli, E. Colombo, P. Cosmina, and G.
Grandi.1995. A putative new peptide synthase operon in Bacillus subtilis: partial characterization. Microbiology 141:645–648.
22. Tsuge, K., T. Ano, and M. Shoda. 1996. Isolation of a gene essential for biosynthesis of the lipopeptide antibiotic plipastatin B1 and surfactin in Bacillus subtilis YB8. Arch. Microbiol. 165:243–251.
23. Ullrich, C., B. Kluge, Z. Palacz, and J. Vater. 1991. Cell-free biosynthesis of surfactin, a cyclic lipopeptide produced by Bacillus subtilis. Biochemistry
30:6503–6508.
24. Umezawa, H., T. Aoyagi, T. Nishikori, A. Okuyama, Y. Yamagishi, M.
Hamada, and T. Takeuchi.1986. Plipastatins: new inhibitors of phospho-lipase A2, produced by Bacillus cereus BMG302-fF67. I. Taxonomy, produc-tion and preliminary characterizaproduc-tion. J. Antibiot. 39:737–744.
25. Vanittanakom, N., and W. Loeffler. 1986. Fengycin—a novel antifungal li-popeptide antibiotic produced by Bacillus subtilis F29-3. J. Antibiot. 39:888– 901.
VOL. 180, 1998 NOTES 1341
at NATIONAL TAIWAN UNIV MED LIB on May 3, 2009
jb.asm.org