利用馬可夫邏輯網路模型與自動化生成的模板加強生醫文獻之語意角色標註 - 政大學術集成
全文
(2) 利用馬可夫邏輯網路模型與自動化生成的模板加強生醫文 獻之語意角色標註 Biomedical Semantic Role Labeling with a Markov Logic Network and Automatically Generated Patterns. 研 究 生:賴柏廷. Student:Po-Ting Lai. 指導教授:蔡宗翰. Advisor:Richard Tzong-Han Tsai 治 政 大 Chao-Lin Liu. 劉昭麟. 立. ‧. ‧ 國. 學 y. sit. io. n. al. er. Nat. 國立政治大學 資訊科學系 碩士論文. Ch. i n U. v. A Thesis submitted to Department of Computer Science National Chengchi University in partial fulfillment of the Requirements for the degree of Master in Computer Science. engchi. 中華民國一百零一年七月 July 2012.
(3) 利用馬可夫邏輯網路模型與自動化生成的模板加強生醫文獻之語 意角色標註. 摘要. 背景: 生醫文獻語意角色標註(Semantic Role Labeling, SRL)是一種自然語. 政 治 大 言處理的技術,其可用來將描述生物過程的語句以 立. predicate-argument. ‧ 國. 學. structures ( PASs ) 表示。SRL 經常受限於 arguments 的 unbalance problem. ‧. 而且需要花費許多的時間和記憶體空間在學習 arguments 之間的相依性。. sit. y. Nat. 方法: 我們提出一 Markov Logic Network ( MLN ) -based SRL 之系統,且此. er. io. 系統使用自動化生成之 SRL 模板同時辨識 constituents 與候選之語意角色。. n. a. v. l CBioProp 語料上來評估。實驗結果顯示我們的方 結果及結論: 我們的方法在 ni. hengchi U. 法勝過目前最先進的系統。此外,使用 SRL 模板後,在時間及記憶體之花 費上亦大幅的減少,而且我們自動化生成之模板亦能幫助建立這些模板。 我們認為本論文提出之方法可以透過增加新的 SRL 模板例如:由生物學家 手動寫的模板,而得到進一步的提升,而且本方法也為於需要處理大量 SRL 語料時,提供一種可能的解法。. i.
(4) Biomedical Semantic Role Labeling with a Markov Logic Network and Automatically Generated Patterns. Abstract. Background: Biomedical semantic role labeling ( SRL ) is a natural language. 政 治 大. processing technique that expresses the sentences that describe biological. 立. processes as predicate-argument structures ( PASs ) .. SRL usually suffers from. ‧ 國. 學. the unbalanced problem of arguments and consuming time and memory on learning the dependencies between the arguments.. ‧. Method: We constructed a Markov Logic Network ( MLN ) -based SRL system,. y. Nat. sit. and the system uses SRL patterns, which utilizes automatically generated. n. al. er. io. approaches, to simultaneously recognize the constituents and candidates of semantic roles.. Ch. i. e. i n U. v. n g c his evaluated on the BioProp corpus. Results and conclusions: Our method The experimental result shows that our method outperforms the state-of-the-art system.. Furthermore, after applying SRL patterns, the costs of the time and. memory are greatly reduced, and our automatically generated patterns are helpful in the development of these patterns.. We consider that our method can. be further improved by adding new SRL patterns such as biological experts manually written patterns and it also provide a possible solution to process large SRL corpus.. ii.
(5) 致謝. 在碩士班兩年的研究生涯中,最需要感謝. 蔡宗翰老師,因為有蔡老師的. 指導才能讓我學習到許多寶貴知識和經驗,以及完成這篇論文。感謝. 劉. 昭麟老師,在政大及 MIG 實驗室對我的照顧,從劉老師學習到許多寶貴知 識及經驗。. 立. 政 治 大. ‧ 國. 學. 在這兩年的時間中,無論在工作或學校中,都受到許多人的指導、照顧 和幫助。首先特別感謝中研院 IASL 實驗室裡的成員,像鴻傑、濟洋、建銘、. ‧. 國豪等學長,以及已經離開 IASL 的岳洋、吉心、晏菁、琮凱等學長姊。還. sit. y. Nat. 有感謝學弟季恩、舟陽、玠鋒等人的幫助。感謝政大 MIG 實驗室裡成員的. er. io. n. al 指導與照顧,像建良學長、怡軒學姊、裕淇學長以及感謝瑞平、家琦、瑋 iv n U engchi 杰、孫暐在許多事情上的幫助。. Ch. 最後感謝我的家人在我的人生與求學生涯上,給我的種種支持,讓我能 夠持續努力下去。 也謝謝口試委員. 許聞廉老師以及宋定懿老師的指導。. 賴柏廷 2012 年 7 月. iii.
(6) TABLE OF CONTENTS CHAPTER 1 Introduction ....................................................................................................... 1 1.1 Background ................................................................................................................. 1 1.2 Biomedical Semantic Role Labeling ( SRL ) ............................................................ 2 1.3 Traditional Formulation of SRL ............................................................................... 3 1.4 Problems ...................................................................................................................... 5 1.4.1 1.4.2. Unbalanced Problem ................................................................................ 5 Dependency Problem ................................................................................ 6. 政 治 大. 1.5 Our Goal ...................................................................................................................... 7. 立. CHAPTER 2 Method ............................................................................................................... 8. Markov Logic Networks .......................................................................... 9. y. Nat. sit. Implement Biomedical Semantic Role Labeling ................................................ 9. n. al. er. Formulating SRL ...................................................................................... 9 Basic formulae ........................................................................................ 10 Conjunction formulae ............................................................................ 11 Global formulae ...................................................................................... 12. io. 2.2.1 2.2.2 2.2.3 2.2.4. Ch. engchi. i n U. v. Patterns for SRL ................................................................................................. 12 2.3.1 2.3.2 2.3.3 2.3.4 2.3.5 2.3.6. 2.4. First-Order Logic ..................................................................................... 8 Markov Networks ..................................................................................... 8. ‧. 2.1.3. 2.3. ‧ 國. Markov Logic ........................................................................................................ 8 2.1.1 2.1.2. 2.2. 學. 2.1. Introduction of the Patterns .................................................................. 12 Tree Pruning............................................................................................ 13 Lexicon Pattern ....................................................................................... 14 Temporal Pattern .................................................................................... 15 Conjunction Pattern ............................................................................... 15 Syntactic Path Pattern ........................................................................... 19. Collective Learning for SRL .............................................................................. 19 2.4.1 2.4.2. Collective Learning ................................................................................ 19 Linguistic Constraints ............................................................................ 19. CHPATER 3 Experiment ....................................................................................................... 21 iv.
(7) 3.1. Dataset ................................................................................................................. 21. 3.2. Experiment Design ............................................................................................. 22 3.2.1 3.2.2. Experiment 1 – The Effect of Automatically Generated Patterns ..... 22 Experiment 2 – Improvement by Using Collective Learning ............. 22. 3.3. Evaluation Metric ............................................................................................... 22. 3.4. t-test ...................................................................................................................... 23. CHAPTER 4 Results and Discussion .................................................................................... 25 4.1. Improvement by Using SRL Patterns ............................................................... 25. 4.2. Improvement by Using Collective Learning .................................................... 26. Biomedical Semantic Role Labeling Corpus ........................................ 28 Biomedical Semantic Role Labeling System ........................................ 28. 學. 4.3.1 4.3.2. ‧ 國. 4.3. 政 治 大 Related Work 立 ...................................................................................................... 28. CHAPTER 5 Conclusion ....................................................................................................... 30. ‧. References................................................................................................................................ 31. n. er. io. sit. y. Nat. al. Ch. engchi. v. i n U. v.
(8) LIST OF FIGURES Figure 1.1: A parsing tree annotated with semantic roles.. ......................................................... 2 Figure 1.2: The pipeline methods of the SRL. ........................................................................... 4 Figure 2.1: The examples show the tree pruning...................................................................... 14 Figure 2.2: The examples for mining association rules. ........................................................... 17 Figure 4.1: An example - ARGM-MNR is failed to be labeled using BIOSMILE + Pattern w/o Auto. ......................................................................................................................................... 26. 立. 政 治 大. ‧. ‧ 國. 學. n. er. io. sit. y. Nat. al. Ch. engchi. vi. i n U. v.
(9) LIST OF TABLES Table 1.1: The statistic of the constituents and semantic roles. .................................................. 6 Table 2.1: The features are used in previous SRL systems. ..................................................... 11 Table 2.2: The information extracted from ARGM-LOC on Figure 2.2.a. .............................. 17 Table 2.3: The transactions are transformed from Figure 2.2................................................... 18 Table 3.1: The statistics on the BioProp corpus. ...................................................................... 21 Table 4.1: The performances of SRL using SRL patterns. ....................................................... 25. 政 治 大 of SRL using collective learning. .............................................. 27 Table 4.3: The performances立 Table 4.2: The distribution of ARGM on BIOSMILE and BIOSMILE + Pattern w/o Auto.... 26. ‧ 國. 學. Table 4.4: The performances of SRL on sentence-wide evaluation metrics. ........................... 27 Table 4.5: The cost of time and memory. ................................................................................. 27. ‧. n. er. io. sit. y. Nat. al. Ch. engchi. vii. i n U. v.
(10) CHAPTER 1 Introduction 1.1 Background The volume of biomedical literature available on the World Wide Web has experienced. 治 processing biomedical literature has 政 Automatically 大. unprecedented growth in recent years.. 立. been receiving lot attentions. Many information extraction ( IE ) researches[1] have shown. ‧ 國. 學. their interested in the challenges of the biomedical text mining.. Because of the difficulties. on processing natural language texts, many biomedical relation-extraction systems only. ‧. Nat. y. However, they frequently In the. sit. consider the main relation targets and the verbs linking them.. Biological processes can. ignore phrases describing location, manner, timing, condition, and extent[2].. n. al. er. io. biomedical field, these modifying phrases are especially important.. i n U. v. be divided into temporal or spatial molecular events, for example activation of a specific. Ch. engchi. protein in a specific cell or inhibition of a gene by a protein at a particular time.. Having. comprehensive information about when, where and how these events occur is essential for identifying the exact functions of proteins and the sequence of biochemical reactions. Detecting the extra modifying information in natural language texts requires semantic analysis tools.. 1.
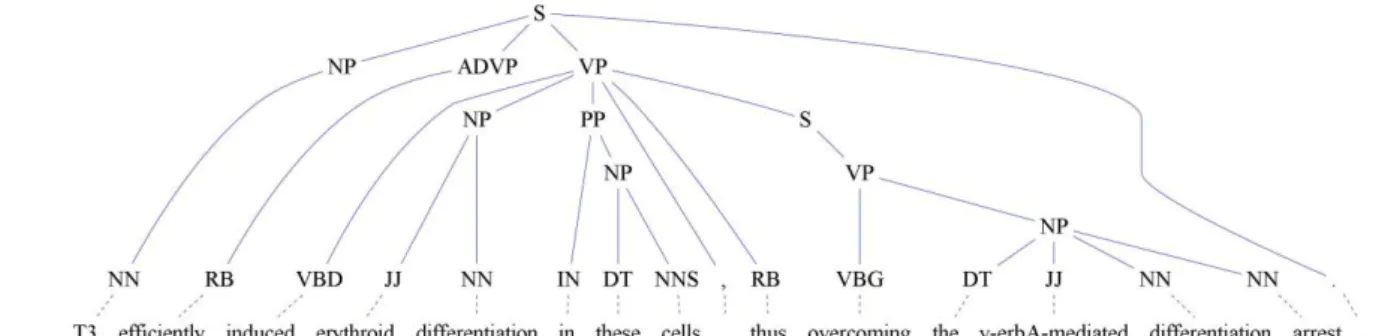
(11) Figure 1.1: A parsing tree annotated with semantic roles.. 立. 政 治 大. ‧ 國. 學. 1.2 Biomedical Semantic Role Labeling ( SRL ). Semantic role labeling ( SRL ) , also called shallow semantic parsing[3], is a popular semantic. ‧. On SRL, sentences are represented by one or more predicate argument Each PAS is composed of a predicate. sit. Nat. structures ( PASs ) , also known as propositions[4].. y. analysis technique.. io. al. er. (e.g., a verb) and several arguments (e.g., noun phrases) that have different semantic roles,. v i n CHere, h ethen term h i U refers to a syntactic constituent of g c argument. n. including main arguments such as an agent and a patient, as well as adjunct arguments, such as time, manner, and location.. the sentence related to the predicate; and the term semantic role refers to the semantic relationship between a predicate (e.g., a verb) and an argument (e.g., a noun phrase) of a sentence. For example, in Figure 1.1, the sentence "IL4 and IL13 receptors activate STAT6, STAT3, and STAT5 proteins in the human B cells." describes a molecular activation process. It can be represented by a PAS in which "activate" is the predicate, "IL4 and IL13 receptors" and "STAT6, STAT3, and STAT5 proteins" comprise the ARG0 and ARG1 respectively, and "in the human B cells" is the location. ARG0 and ARG1 have different defines on different. 2.
(12) predicate, and describe agent and patient respectively.. Thus, the agent, patient, and location. are the arguments of the predicate.. 1.3 Traditional Formulation of SRL SRL has being formulated as a classification problem in which supervised machine learning methods can be applied [2, 5, 6].. In order to constructing such a classifier, it is essential to. select an annotated corpus and a PAS standard, such as PropBank[7]. most general formulation of SRL, a pipeline method [8].. Figure 1.2 shows the. As shown in Figure 1.2.a, SRL. 政 治 大. usually starts with accepting a syntactic structure of a sentence (parse tree), because the. 立. The second step (Figure 1.2.b). structure encode more information such as the headwords [9].. This step can be achieved. by using a part-of-speech (POS) tagger with some filtering rules.. Figure 1.2.c and Figure. ‧. ‧ 國. 學. is predicate identification that identifies the verb on the sentence.. 1.2.d shows the two major tasks of SRL. Figure 1.2.c is called argument identification that. Nat. n. al. Figure 1.2.d refers to as the argument. er. io. syntactic tree, contain semantic roles or not.. sit. y. identifies the word boundaries by determining whether the constituents, the nodes on the. i n U. v. classification step that assigns appropriate semantic role labels to the constituents. Finally,. Ch. engchi. Figure 1.2.e checks whether the semantic roles are legal or not[10] by checking constraints such as word boundaries cannot be overlapped and determines the final semantic roles of the sentence. This step has been shown as an importance step in SRL [10]. Some systems, such as BIOSMILE[2], treat argument identification and argument classification as a single step, and recognize word boundaries and semantic roles simultaneously.. However, the formulation usually suffers from the unbalanced problem of. semantic role labels, because there are many kinds of semantic role labels for instance BioProp defines thirty-two kinds of semantic role labels. Furthermore, SRL is a non-i.i.d (individual and identical distribution) problem. Following the formulation, the dependencies 3.
(13) between semantic roles could not be known immediately until the last step.. In the next. section, we will describe our observations on both problems on the current biomedical SRL approaches.. a.. Figure 1.2: The pipeline methods of the SRL. The input of SRL is the syntactic tree of the sentence, and the nodes of the syntactic tree are called constituents.. 立. Predicate identification identifies the predicate constituent.. n. al. er. io. sit. y. Nat. c.. ‧. ‧ 國. 學. b.. 政 治 大. Ch. engchi. i n U. v. Argument identification identifies the argument constituents.. 4.
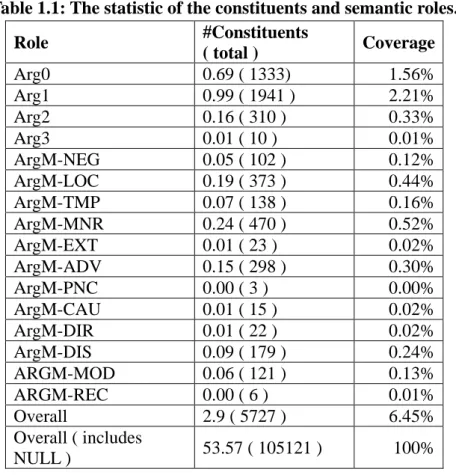
(14) d.. Argument classification assigns appropriate the semantic role labels to the constituents.. 立. ‧ 國. 學. Checking whether the semantic role labels violate the linguistic constraints.. ‧. io. sit. y. Nat. n. al. er. e.. 政 治 大. Ch. engchi. i n U. v. 1.4 Problems 1.4.1 Unbalanced Problem At first, we found the major reason causing unbalanced semantic roles comes from the stages of argument identification and argument classification.. Table 1.1 shows our statistics of the. constituents that possess semantic role labels on BioProp.. As shown in Table 1.1, about. 93.55% of the constituents in the syntactic tree do not have any semantic role label, meaning 5.
(15) that these constituents are labeled with the NULL label in the dataset. By further analyzing these semantic roles, we observed that some of them can be found by simple patterns such as their syntactic path.. For instance, in Figure 1.2, the inducer “T3” can be recognized by the. syntactic path from the constituent to the verb “NP>S<VP<VBD”.. However, most systems. take all constituents as the input, and spend a lot time on tuning the weights for features of the NULL label and ignore the semantic roles with few instances. Table 1.1: The statistic of the constituents and semantic roles. #Constituents Role Coverage ( total ) Arg0 0.69 ( 1333) 1.56% Arg1 0.99 ( 1941 ) 2.21% Arg2 0.16 ( 310 ) 0.33% Arg3 0.01 ( 10 ) 0.01% ArgM-NEG 0.05 ( 102 ) 0.12% ArgM-LOC 0.19 ( 373 ) 0.44% ArgM-TMP 0.07 ( 138 ) 0.16% ArgM-MNR 0.24 ( 470 ) 0.52% ArgM-EXT 0.01 ( 23 ) 0.02% ArgM-ADV 0.15 ( 298 ) 0.30% ArgM-PNC 0.00 ( 3 ) 0.00% ArgM-CAU 0.01 ( 15 ) 0.02% ArgM-DIR 0.01 ( 22 ) 0.02% ArgM-DIS 0.09 ( 179 ) 0.24% ARGM-MOD 0.06 ( 121 ) 0.13% ARGM-REC 0.00 ( 6 ) 0.01% Overall 2.9 ( 5727 ) 6.45% Overall ( includes 53.57 ( 105121 ) 100% NULL ). 立. 政 治 大. ‧. ‧ 國. 學. n. er. io. sit. y. Nat. al. Ch. engchi. i n U. v. 1.4.2 Dependency Problem Based on our error analysis on the output generated by BIOSMILE, some sentences failed to be expressed as PAS, which mainly results from incorrect SRL due to the complexity of the sentences. An example is shown in Figure 1.2.e.. If we have enough knowledge of the. linguistic constraints between semantic roles, it might help us in assigning the appropriate semantic role labels. This is also called collective learning.. 6. We think this observation.
(16) emphasizes the importance of collectively learned semantic roles.. 1.5 Our Goal In this paper, we focus on 1) how to automatically select and assign initial semantic roles; 2) how to enhance SRL with collectively learned semantic roles through using a Markov Logic Network[11]. method.. Following is an overview of this paper.. In Chapter 2, we describe our. The subsection 2.1 introduces MLN proposed by [12].. SRL system is described in subsection 2.2.. The implementation of our. The subsection 2.3 proposes the method of using. Due to the difficulties for 政 治 大 employing biological experts in manually writing the SRL patterns, we propose an automatic 立 SRL patterns to select and assign initial semantic role labels.. pattern generation method.. In the subsection 2.4, we introduce the collectively learned. ‧ 國. 學. semantic roles method that implements linguistic constraints. In Chapter 3, we detail the. io. sit. y. Finally, Chapter 5 concludes the contributions of. n. al. er. this paper.. Nat. experimental results and related analysis.. In Chapter 4, we show the. ‧. experiments designed for examining the effect of our methods.. Ch. engchi. 7. i n U. v.
(17) CHAPTER 2 Method 2.1 Markov Logic 2.1.1. First-Order Logic. 政 治 大 In FOL, formulae consist of four types of symbols: constants, 立 variables, functions, and predicates. Constants represent MLN combines first order logic (FOL) and Markov networks.. ‧ 國. 學. objects in a specific domain ( e.g. Part-of-speech: NN, VB, etc. ) .. Variable is the range over. the objects ( e.g., PoS ( Part-of-speech ) , where PoS { pos | pos Part of speech} ) .. ‧. Functions represent mappings from tuples of objects to objects ( e.g., ChildrenOf , where. sit. y. Nat. ChildrenOf (i) children of tree node i ) . Predicates represent relationships among objects. n. al. may belong to specific types.. er. io. ( e.g., PoS of headword ) , or attributes of objects ( e.g., Arg0 ) . Constants and variables. i n U. v. An atom is a predicate symbol applied to a list of arguments,. Ch. engchi. which may be constants or variables ( e.g., role ( p, i, r ) ) . whose. arguments. are. all. A ground atom is an atom. constants. (. e.g.,. event _ trigger { p | p selected 30 frequent verbs} ) . A world is an assignment of truth values to all possible ground atoms.. A knowledge base ( KB ) is a partial specification of a. world; each atom in it is true, false or unknown.. 2.1.2 A. Markov Networks Markov. network. represents. the. joint. distribution. variables X {X 1 ,, X n } X as a product of factors: P X x . of. a. set. of. 1 f k xk , where each Z k. factor f k is a non-negative function of a subset of the variables x k , and Z is a normalization 8.
(18) constant.. The distribution is usually equivalently represented as a log-linear form:. P X x . 1 exp wi g j x , where the features g i x are arbitrary functions of ( a subset Z i . of ) the variables’ states.. 2.1.3. Markov Logic Networks. An MLN is a set of weighted first-order formulae. Together with a set of constants representing objects in the domain, it defines a Markov network with one variable per ground distribution over possible worlds 政 The治probability 大 . atom and one feature per ground formula.. 立. 1 exp wi g j x where Z is the partition function, is the set of Z iF jGi . 學. ‧ 國. is given by P X x . all first-order formulae in the MLN, g j is the set of groundings of the i-th first-order formula,. ‧. and g j x 1 if the j-th ground formula is true and g j x 0 otherwise.. Nat. y. Markov logic General. er. io. sit. enables us to compactly represent complex models in non-i.i.d. domains.. algorithms for inference and learning in Markov logic are discussed in Richardson and. al. n Domingos[13].. Ch. i n U. v. We uses 1-best MIRA online learning method [14] for learning weights and. engchi. employs cutting plane inference [15] with integer linear programming as its base solver for inference at test time as well as during the MIRA online learning process.. To avoid the. ambiguity between the predicates in FOL and SRL, we refer the predicate in SRL as “event trigger” from now on.. 2.2 Implement Biomedical Semantic Role Labeling 2.2.1. Formulating SRL. Our SRL system incorporates three components: (1) SRL patterns; (2) collective learning formulae; (3) a MLN-based classifier. SRL patterns: The SRL patterns are the patterns described in subsection 2.3. 9. We use.
(19) pattern _ match ( p, i, r ) to describe that there is an event trigger p and the constituent i has a semantic role r. Collective learning formulae: The collective learning formulae are the formulae described in subsection 2.4. MLN-based classifier: Our MLN-based classifier uses the features of BIOSMILE, transform them into the formulae.. In subsection 2.2.2, we will propose our method about. how to transform these features into the formulae and how to incorporate SRL patterns and. 政 治 大 formulae using only annotated PAS information. In section 2.2.4, we further apply the 立. the classifier.. In section 2.2.3, we propose a method to automatically generate conjunction. collective learning on SRL.. ‧ 國. 學. In our formulation, we use event _ trigger ( p) to express a constituent p that is an. ‧. event trigger; event _ type ( p, t ) to express that the event type of p is t such binding;. y. al. n. Basic formulae. sit. io. 2.2.2. er. semantic role r .. Nat. role ( p, i, r ) to express that there is an event trigger p and a constituent i with the. Ch. i n U. v. Basic formulae are derived from the features used on the SRL systems[5, 16-18] that are. engchi. based on Maximum Entropy Model ( ME ), Support Vector Machine ( SVM ). Table 2.1. These features are also used on BIOSMILE. their contributions on SRL.. As shown in. These features have been proved. To apply these features on our classifier, we transform the. features into the formulae, since MLN only accepts the formulae rather than features. A basic formula consists of two predicates, one corresponding to the event trigger and the other one is a feature of a constituent.. For example, the headword feature could be. expressed in FOL as pattern _ match ( p, i, r ) headword ( i, w ) role ( p, i, r ) , where w is the headword of the constituent i .. If the “+” symbol appears before a variable, it. indicates that each different value of the variable has its own weight. 10.
(20) al. n. . io. . Nat. . ‧. . 立. 學. . 政 治 大. y. . sit. . er. . ‧ 國. . Table 2.1: The features are used in previous SRL systems. BASIC FEATURES Predicate – The predicate lemma Path – The syntactic path through the parsing tree from the constituent being classified to the predicate Constituent type Position – Whether the phrase is located before or after the predicate Voice – passive if the predicate has a POS tag VBN, and its chunk is not a VP, or it is preceded by a form of "to be" or "to get" within its chunk; otherwise, it is active Head word – Calculated using the head word table described by Collins (1999) Head POS – The POS of the Head Word Sub-categorization – The phrase structure rule that expands the predicate's parent node in the parsing tree First and last Word and their POS tags Level – The level in the parsing tree PREDICATE FEATURES Predicate's verb class Predicate POS tag Predicate frequency Predicate's context POS Number of predicates FULL PARSING FEATURES Parent, left sibling, and right sibling paths, constituent types, positions, head words, and head POS tags Head of Prepositional Phrase (PP) parent – If the parent is a PP, then the head of this PP is also used as a feature COMBINATION FEATURES Predicate distance combination Predicate phrase type combination Head word and predicate combination Voice position combination OTHERS Syntactic frame of predicate/NP Headword suffixes of lengths 2, 3, and 4 Number of words in the phrase Context words & POS tags. 2.2.3. Ch. engchi. i n U. v. Conjunction formulae. In addition to the basic formulae described above, we also employ conjunction formulae. We use a similar approach described in the subsection 2.3 to generate conjunction formulae. However, unlike those patterns would like to achieve a higher recall and not care about the precision, the conjunction formulae should as possible as improving the recall and precision. Therefore we use Apriori algorithm to generate conjunction formulae. 11.
(21) Apriori algorithm has been described in the subsection 2.3, and to generate conjunction formulae we set the default minimum support and confidence are 15 times and 80%, and we believe the values could generate the reliable conjunction formulae. Conjunction formulae are composed of three or more predicates, one is the event trigger and the others are the linguistic properties of a constituent. For instance,. event _ trigger ( p) constituen t _ type (i, " PP") firstword (i, " in" ) lastword (i, " cell" ) role ( p, i , " ARGM - LOC" ) means that the constituent i should be labeled as “ARGM-LOC” when its constituent type is. 政 治 大. “PP”, its first word is “in”, and its last word is “cell”.. 2.2.4. 立. Global formulae. ‧ 國. 學. Basic formulae and conjunction formulae are local formulae whose conditions only consider the observe predicates. That is the dependencies of the semantic roles do not take into. ‧. considered. The global formula is the condition of the formula including hidden predicates. sit. y. In our system the hidden predicate is. We use the global formulae described in subsection 2.4 to the collectively. io. al. er. role ( p, i , r ) .. Nat. or the constraints that cannot be violated.. n. learned semantic roles with dependences including tree collective and path collective.. 2.3 Patterns for SRL 2.3.1. Ch. engchi. i n U. v. Introduction of the Patterns. In this section, we propose the formal definition of our patterns. In ideal situation, the patterns of SRL can express the sentences as PASs. biological experts.. However, it is difficult without the help of the. For example, a pattern indicates that the noun phrase that appears in. front of an active verb such as bind is the agent, but another pattern indicates that the protein before bind is the agent. It is difficult to determine which pattern is corrected.. The first. pattern might be wrong while the noun phrase describes the process about a protein. However the second pattern might be fail while the protein could not be recognized. Therefore, it requires to manually design the dependencies of the patterns. 12. Because it is.
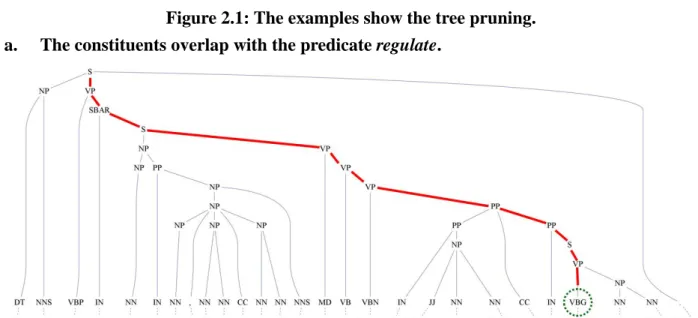
(22) difficult for human to manually design the dependencies.. Our patterns are designed to. answer what are the candidate labels of the semantic roles on the sentences rather than what are the appropriate semantic role labels.. Our patterns focus on removing the constituents. that should not be assigned semantic role labels and recognizing the candidate labels of the semantic roles.. 2.3.2. Following sections describe our SRL patterns.. Tree Pruning. Since the ratio of the constituents with semantic roles is much lower than all constituents. Some SRL systems 政 治 大 also use the pruning methods[17] or pre/post-processing filtering method[10] on the SRL to 立 The goal of tree pruning is to reduce the number of the constituents.. reduce the complexity or improve the performance.. These methods are also used in our SRL. ‧ 國. 學. patterns. We use two different tree pruning methods in our SRL.. If a constituent overlaps with. the. Removing these overlapped. sit. y. Nat. predicate and it should not be assigned the semantic role.. ‧. the constituents in the same path with the predicate.. The first one is removing. io. roles but also make training/testing efficiently.. n. al. Ch. er. constituents before classification not only make sure they cannot be assigned the semantic Figure 2.1.a shows an example.. n U engchi. iv. The. second one is that SRL prefer to annotate semantic roles on the phrase rather than the token, while the constituents 1) are leaves, 2) do not have any sibling 3) are stop words, they should be removed.. Figure 2.1.b shows an example.. 13.
(23) 立. The constituents are stop words or without the siblings.. 學 ‧. io. sit. y. Nat. n. al. er. b.. 政 治 大. ‧ 國. a.. Figure 2.1: The examples show the tree pruning. The constituents overlap with the predicate regulate.. 2.3.3. Lexicon Pattern. Ch. engchi. i n U. v. Lexicon pattern assigns the semantic role label to the constituent. systems use lexicon features on argument identification/classification.. Like most of the SRL Here we describe the. semantic role labels could be found by their words, and we use the string match method to identify these semantic role labels. Discourse ( ARGM-DIS ) : Discourse connects a sentence to a preceding sentence, it is not necessary to use classification to find them but a simple list. Modals ( ARGM-MOD ) , Negation ( ARGM-NEG ): While the predicate next to these words, we would assign the words the semantic roles. role pairs with the words list. 14. We collect these words and semantic.
(24) 2.3.4. Temporal Pattern. The semantic roles with number or time are difficult to be recognized. These semantic roles also make the sentences much complex.. Recognizing these semantic roles before. classification could decrease the complexity of the sentences.. We manually write the. patterns to recognize these semantic roles Extent Marker ( ARGM-EXP ) : Extent marker indicates the amount of change occurring from an action such as “… fold”.. We observe that extent markers usually are the siblings of. the verb. Therefore, we design our pattern as following: if there is a trigger of the extent. 治 政 markers such as “%” or “fold”, the constituent of the sibling 大 of the verb which contains this 立 trigger would be assigned the extent marker. ‧ 國. 學. Temporal Marker ( ARGM-TMP ) : Temporal marker indicates when an action took Like extent marker, temporal markers usually are the siblings of the verb.. ‧. place.. we use the same methods to find temporal markers.. Therefore,. Furthermore, temporal markers. y. Nat. io. sit. sometimes appear in the head of sentence, we also assign the temporal marker to the. n. al. er. constituent which has the trigger of temporal marker such as “hour” and is the start of the sentence.. 2.3.5. Ch. Conjunction Pattern. engchi. i n U. v. In addition to all above patterns, there still have a lot of potential patterns could be used to annotate the semantic roles. Here we propose a method that uses the association rule mining and can automatically generate the patterns that conjunct several features of the constituents. For instance, first_word ( i, “in” ) ˄ last_word ( i , “cell” ) => role ( p, i, “ARGM-LOC” ), this pattern means the constituent i started with “in” and ended with “cell” should be assigned the locative modifiers ARGM-LOC.. In subsection 2.3.5.1, we introduce the association rule. mining; in subsection 2.3.5.2, we propose our formulation of the transactions on SRL; in subsection 2.3.5.3, we describe our filtering methods to select the conjunction patterns. 15.
(25) 2.3.5.1 Association Rule Mining Association rule mining[19] is to discover the interesting relations, called association rules, from certain database, and it also is a popular research method.. An association rule is a rule. like “If a person buys wine and bread, he/she often buys cheese, too”. SRL patterns are very similar with the association rules.. We found that the. For an instance, a SRL pattern can. be written as a rule like “If a constituent starts with the word in and ends with the word cell, it often plays an ARGM-LOC”. conjunction patterns.. Therefore, we apply association rule mining to generate. In order to discover the interesting relations, it is necessary to define. 政 治 大. four things including item, transaction, support and confidence.. 立. An item is the object. participating in the rules, continuing the example, the started word in, the ended word cell and. ‧ 國. 學. the semantic role ARGM-LOC are the items. The transaction is a collection of the items. The support is the number of the itemset, a subset of the transaction, appearing in the. ‧. A minimum support could be used to make sure that the. y. Nat. The confidence is the number of the rule hold. io. divided by the number of the condition hold.. A minimum confidence could make sure that. n. al. sit. mined rules are not to overfit the database.. er. collection of the transactions.. mined rules often are corrected in the database.. Ch. i n U. v. A maximum confidence could make sure. engchi. that mined rules are not obviously in the database.. In our paper, we will focus on how to. discover the rules instead of how to implement the association rule mining method. 2.3.5.2 Formulate the Transaction By observing the individual semantic role, we find sometimes the semantic role could be determined by its first and last words such as a phrase likes “in…cell” usually play a role ARGM-LOC.. Therefore, we propose a method which could automatically generate the. patterns like that, and the steps are below:. 16.
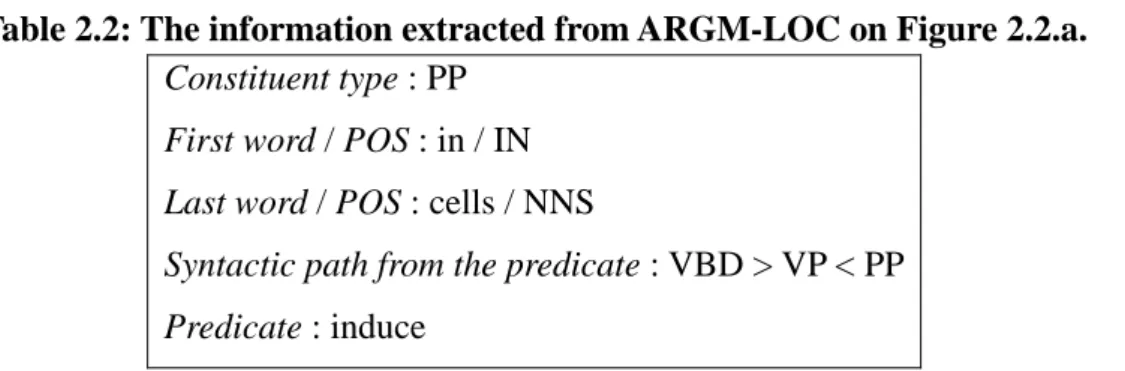
(26) Figure 2.2: The examples for mining association rules. a. T3 efficiently induced erythroid differentiation in these cells, thus overcoming the v-erbA-mediated differentiation arrest.. 政 治 大. b. In contract mRNA representing pAT 591/EGR2 was not expressed in these cells.. 立. ‧. ‧ 國. 學. n. er. io. sit. y. Nat. al. i n U. v. Step 1: Extracts the information about all the arguments include constituent type, first. Ch. engchi. word and last word, syntactic path from the predicate and the predicate. information as the items.. We treat these. For instance, in Figure 2.2.a, for ARGM-LOC, we could extract. the information as following: Table 2.2: The information extracted from ARGM-LOC on Figure 2.2.a. Constituent type : PP First word / POS : in / IN Last word / POS : cells / NNS Syntactic path from the predicate : VBD > VP < PP Predicate : induce Step 2: We treat each constituent with the semantic role as a transaction, and its 17.
(27) information extracted in Step 1 is its items.. For instance, we have two sentences as shown in. Figure 2.2, and we could transform them into the transactions:. Table 2.3: The transactions are transformed from Figure 2.2. Itemset FW ( T3 ) , LW ( T3 ) , CT ( NP ) , PATH ( NP>S<VP<VBD ) , event_trigger ( induce ) , ROLE ( ARG0 ) FW ( efficiently ) , LW ( efficiently ) , CT ( ADVP ) , PATH ( ADVP>S<VP<VBD ) , event_trigger ( induce ) , ROLE ( ARGM-MNR ) FW ( erythroid ) , LW ( differentiation ) , CT ( NP ) , PATH ( VBD>VP<NP ) , event_trigger ( induce ) , ROLE ( ARG1 ) FW ( in ) , LW ( cell ) , CT ( PP ) , PATH (VBD>VP >S<PP ) , event_trigger ( induce ) , ROLE ( ARGM-LOC ). 政 治 大 FW ( overcoming ) , LW ( arrest ) , CT ( S ) , PATH ( VBD>VP<S ) , event_trigger ( induce ) , ROLE ( ARGM-ADV ) 立 FW ( in ) , LW ( contrast ) , CT ( PP ) , PATH ( PP>S<VP<VP<VBN ) , event_trigger ( express ) , ROLE ( ARGM-DIS ) FW ( thus ) , LW ( thus ) , CT ( RB ) , PATH ( VBD>VP<RB ) , event_trigger ( induce ) , ROLE ( ARGM-DIS ). ‧ 國. 學. FW ( mRNA ) , LW ( 591/egr2 ) , CT ( NP ) , PATH ( NP>S<VP<VBN ) , event_trigger ( express ) , ROLE ( ARG1 ) FW ( not ) , LW ( not ) , CT ( RB ) , PATH ( RB>VP<VBD ) , event_trigger ( express ), ROLE ( ARGM-NEG ) FW ( in ) , LW ( cell ) , CT ( PP ) , PATH ( VBD>VP<S<PP ) , event_trigger ( express ), ROLE ( ARGM-LOC ). ‧ y. Nat. sit. Step 3: Using association rule mining, we could generate the rule likes. n. al. er. io. event _ trigger ( p) constituen t _ type (i, " PP") firstword (i, " in" ) lastword (i, " cells" ) lastword _ POS (i, " NNS" ) role ( p, i , " ARGM - LOC" ) 2.3.5.3 Select the Patterns. Ch. engchi. i n U. v. However, the patterns generated in Step 3 probably are not suite for the SRL.. We observe. the characteristics of different semantic roles, and we apply following the metrics to select SRL patterns: a) The conjunction patterns for ARGX must contain “the syntactic path and the predicate type” and must appear more than 2 times, and the condition should only include these two items. b) The conjunction pattern for ARGM-LOC should contain either “the first word and the last word” or “the first word and the syntactic path” and should appear more than 18.
(28) 2 times. c) The conjunction pattern for the other ARGM should contain “the word and the syntactic path” and should appear more than 2 times.. 2.3.6. Syntactic Path Pattern. In addition to all above methods, we use the shortest syntactic path patterns, while the constituents have no candidate semantic role label, we check whether the constituent has similar syntactic path with semantic roles that appear in training set, if it exists, the. 政 治 大 2.4 Collective Learning 立for SRL constituent would be assigned the semantic role label.. Collective Learning. ‧ 國. 學. 2.4.1. Collective learning is also known as collective classification.. For instance, the disease-gene related. ‧. they assign appropriate labels to the instances.. In classification problems,. y. sit. In this problem, it assumes whether the document is disease-gene related or not,. io. that is independent with other reference documents.. al. However, there is rich information on. v i n UsingC collective learning can h e n g c h i Ubenefit from this information.. n. its reference documents.. er. documents.. Nat. document classification problem distinguish disease-gene related document from other. And. MLN also show that it performs well on collection learning[20].. 2.4.2. Linguistic Constraints. The linguistic constraints[10] have shown their contributions on SRL.. In our paper, we. called the linguistic constraints as tree collective and path collective. Tree collective indicates that two or more arguments in a sentence may be assigned the same semantic role, which contradicts PAS.. To prevent this, we use the formula. event _ trigger ( p) core_arg (r ) role ( p, i, r ) 1 This formula ensures that each semantic role is assigned to only one constituent.. 19. We called.
(29) the formula as tree collective, since the formula limits an event trigger could not has more than one core argument ( the number argument : ARGX ). Furthermore, the arguments may overlap when a node and it antecedent node(s) are all assigned semantic roles.. The formula. overlap (i, j ) role ( p, i, r1 ) role ( p, j, r2 ) 0. ensures that if two or more constituents overlap, then only one can be assigned a semantic role.. We called the formula as path collective, since the formula limits the argument could. not appear in the same path on the syntactic tree.. 立. 政 治 大. ‧. ‧ 國. 學. n. er. io. sit. y. Nat. al. Ch. engchi. 20. i n U. v.
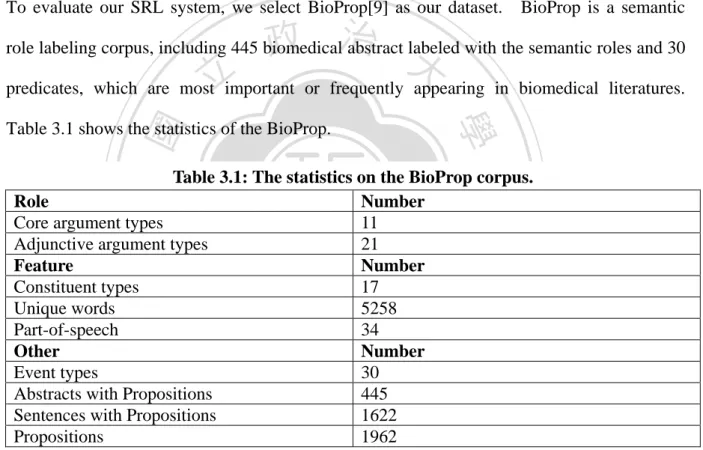
(30) CHPATER 3 Experiment 3.1 Dataset To evaluate our SRL system, we select BioProp[9] as our dataset.. 政 治 大. BioProp is a semantic. role labeling corpus, including 445 biomedical abstract labeled with the semantic roles and 30. 立. predicates, which are most important or frequently appearing in biomedical literatures.. ‧ 國. 學. Table 3.1 shows the statistics of the BioProp.. ‧. Table 3.1: The statistics on the BioProp corpus. Role Number Core argument types 11 Adjunctive argument types 21 Feature Number Constituent types 17 Unique words 5258 Part-of-speech 34 Other Number Event types 30 Abstracts with Propositions 445 Sentences with Propositions 1622 Propositions 1962. n. er. io. sit. y. Nat. al. Ch. engchi. i n U. v. Core arguments play the major semantic role of the event, including ARGX, R-ARGX and C-ARGX. ARGX usually plays the agent, patients and objects; R-ARGX indicates the start of the clause that describes ARGX; C-ARGX describes the continuous ARGX. Adjunctive arguments play the location, manner, temporal, extent used to indicate the state of the event.. 21.
(31) 3.2 Experiment Design 3.2.1. Experiment 1 – The Effect of Automatically Generated Patterns. In this experiment, we evaluate the effect of using SRL patterns. effect of automatically generated patterns.. In order to evaluate the. We compare three different configurations.. 1). BIOSMILE : This only implements the basic formulae but with slightly difference is that. event_trigger ( p ) replaces pattern _ match ( p, i, r ) , which means the patterns are not used.. 2) BIOSMILE + pattern w/o auto : To examine the effect of the patterns, another. basic 治formulae. In this configuration, 政 pattern _ match _ wo _ auto _ gen ( p, i, r ) replaces 大 pattern _ match ( p, i, r ) , which 立. configuration. implements. the. means automatically generated patterns are not used in this configuration.. 3) BIOSMILE +. ‧ 國. 學. pattern : The configuration implements all the patterns and the formulae including basic. ‧. formulae and conjunction formulae. Comparing configuration 2 and configuration 3 could. io. y. sit. Experiment 2 – Improvement by Using Collective Learning. er. 3.2.2. Nat. show the effects of using automatically generated patterns.. In this experiment, we examine whether the patterns incorporated with collective learning. n. al. could further enhance SRL.. v i n C compare We different configurations. U h e n gfour i h c. 1) BIOSMILE :. BIOSMILE is the same configuration with that is used in experiment 1. 2) BIOSMILE + pattern : it is also the same with that is used in experiment 1.. 3) BIOSMILE + CL :. BIOSMILE incorporate with the collective learning. 4) BIOSMILE + pattern + CL : BIOSMILE + pattern incorporate with the collective learning.. 3.3 Evaluation Metric The results are given as F-score using the CoNLL-05[8] evaluation script and defined as F. 2 P R , where P denotes the precision and R denotes the recall. PR. for calculating precision and recall are as follows:. 22. The formulae.
(32) Precision . Reacll . the number of correctly recognized arguments the number of recognized arguments. the number of correctly recognized arguments the number of true arguments. Furthermore, we also evaluate the F-score with sentence-level Fs , which we denote. Ps and Rs as follows: Precision . the number of correctly recognized Propositions the number of recognized Propositions. Reacll . 3.4 t-test. the number of correctly PASs the number of true PASs. 政 治 大. 立. In order to develop a much fairer environment, we apply a two-sample paired t-test, which is. ‧ 國. 學. defined as following:. ‧. The null hypothesis, which states that there is no difference between the two. y. Nat. configurations A and B, is given as. io. sit. H 0 : A B. n. al. er. where A is the true mean F-score of configuration A, B is the mean of the configuration B, and the alternative hypothesis is. Ch. engchi. i n U. v. H1 : A B. A two-sample paired t-test is applied since we assume the samples are independent.. As. the number of samples is large and the samples’ standard deviations are known, the following two-sample t-test is suitable: t. (X A X B ) S A2 S B2 n A nB. If the resulting t-score is equal to or less than 1.67 with a degree of freedom of 29 and a statistical significance level of 95%, the null hypothesis is accepted; otherwise it is rejected. 23.
(33) To get the average F-scores and their deviations required for the t-test, we randomly sampled thirty training sets (g1, ..., g30) and thirty test sets (d1, ..., d30) from the 500 abstracts. We trained the model on gi and tested it on di. Afterwards, we summed the scores for all thirty test sets, and calculated the averages for performance comparison.. 立. 政 治 大. ‧. ‧ 國. 學. n. er. io. sit. y. Nat. al. Ch. engchi. 24. i n U. v.
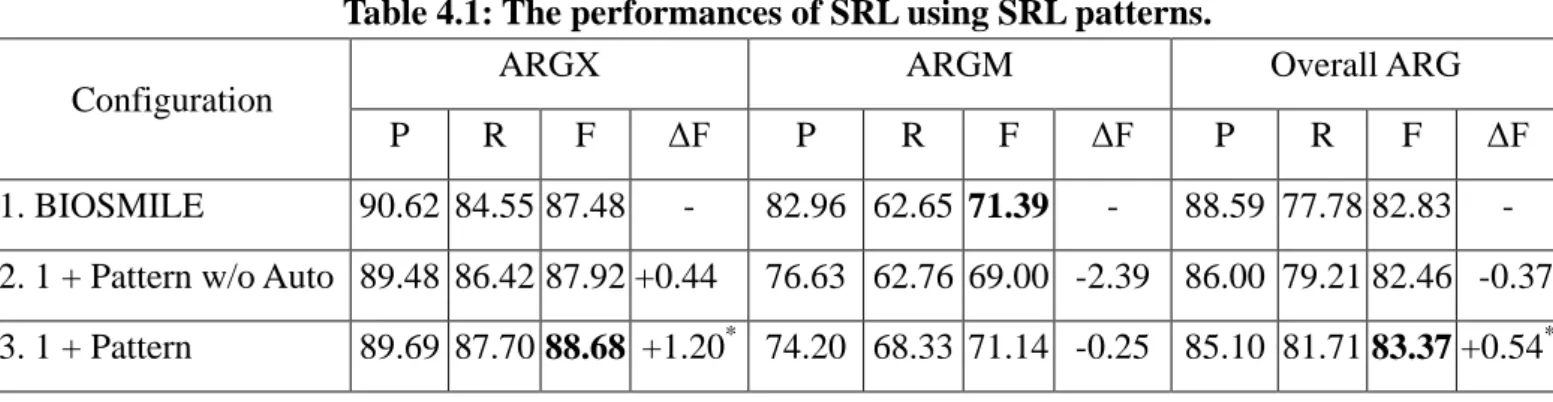
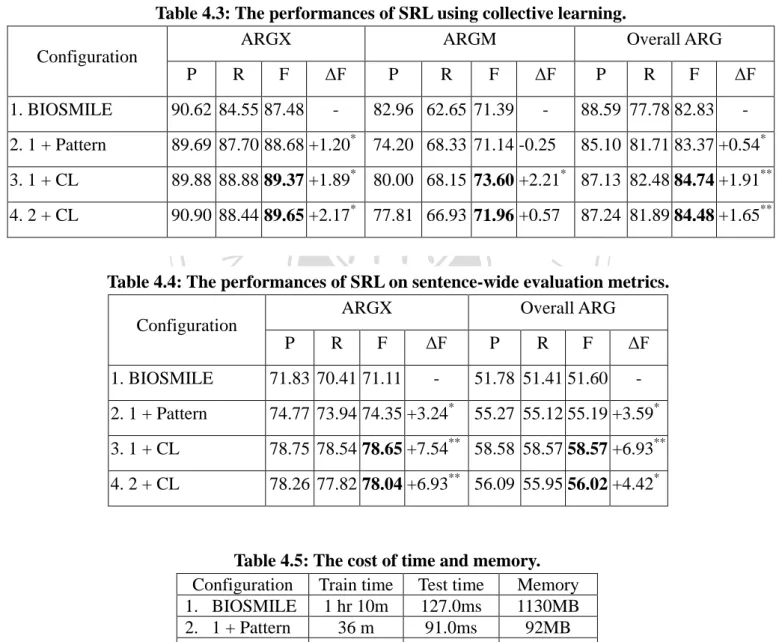
(34) CHAPTER 4 Results and Discussion 4.1 Improvement by Using SRL Patterns Table 4.1 shows the results of using SRL patterns on CoNLL evaluation metrics. three different configurations.. 政 治 大. There are. First, the config. 1 is BIOSMILE. Secondly, the config. 2 is. 立. SRL patterns excluded automatically generated patterns and basic formulae.. ‧ 國. 學. is SRL patterns, basic formulae and automatically generated formulae.. The config. 3 We use ‘*’ to. indicate the configuration has statistically significant improvement with the config. 1 and 2.. ‧. The results show that the config. 2 outperforms the BIOSMILE by 0.44% on ARGX, and. y. Nat. er. io. sit. the config. 3 outperforms the BIOSMILE by 1.20% and 0.54% on ARGX and Overall ARG, respectively. The config. 2 and 3 decrease 2.39% and 0.25% on ARGM respectively.. al. n. v i n C config. 2 and U As show in Table 4.1 that bothhthe e n g c h i 3 perform better in recall but worse on. precision. The reason might be that lacks negative examples that SRL patterns remove the constituents which were considered to be without semantic role labels. Table 4.1: The performances of SRL using SRL patterns. ARGX ARGM Configuration P 1. BIOSMILE. R. F. 90.62 84.55 87.48. ΔF -. 2. 1 + Pattern w/o Auto 89.48 86.42 87.92 +0.44 3. 1 + Pattern. P. R. F. 82.96 62.65 71.39. ΔF -. Overall ARG P. R. F. 88.59 77.78 82.83. ΔF -. 76.63 62.76 69.00 -2.39 86.00 79.21 82.46 -0.37. 89.69 87.70 88.68 +1.20* 74.20 68.33 71.14 -0.25 85.10 81.71 83.37 +0.54*. 25.
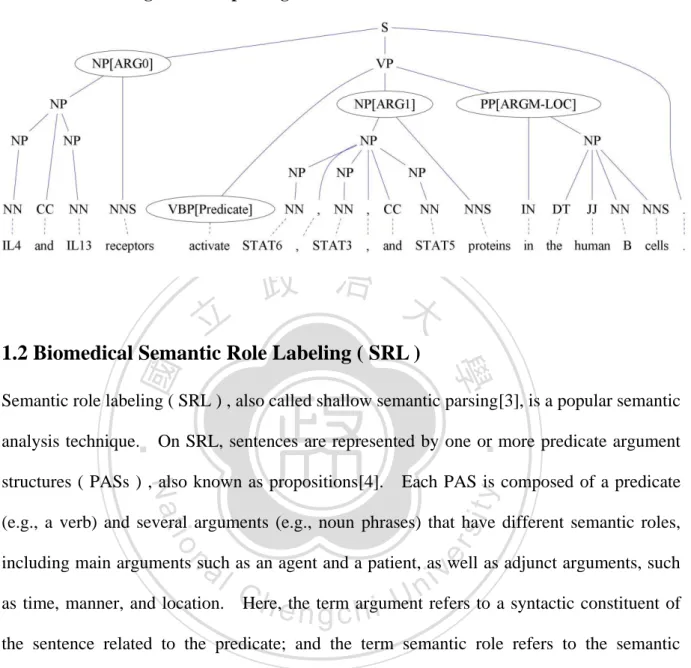
(35) Table 4.2: The distribution of ARGM on BIOSMILE and BIOSMILE + Pattern w/o Auto. BIOSMILE BIOSMILE + Pattern w/o Auto ARGM corr. excess missed corr. excess missed ARGM-ADV 639 332 1102 706 552 933 ARGM-LOC 1465 364 797 1546 494 693 ARGM-MNR 1952 341 811 2025 527 671 ARGM-MOD 696 11 49 709 29 36 ARGM-NEG 521 5 43 536 48 28 ARGM-TMP 286 93 527 327 140 483 Figure 4.1: An example: ARGM-MNR is failed to be labeled using BIOSMILE + Pattern w/o Auto. Collectively , these data suggest that [NFATARG1] [canARGM-MOD] be [activatedEvent Trigger] and IL-2 can be produced [in a calcineurin independent mannerARGM-MNR]. 政 治 大 improve on ARGX, but decrease 立 on ARGM. Table 4.2 shows the individual distribution of Furthermore, we analysis the possible reasons why the config. 2 uses the patterns could. ‧ 國. After applying the patterns, the both number of corrected and. excessed semantic roles on the ARGM increased.. 學. different semantic roles.. Figure 4.1 shows that the patterns of the. ‧. config. 2 without automatically generated patterns are hardly on recognizing the ARGM such. sit. y. Nat. as ARGM-MNR which needs the other information such as the last word, the constituent. n. a l Collective Learning 4.2 Improvement by Using Ch. engchi. er. io. started with “in” and ended with “manner” usually indicates ARGM-MNR.. i n U. v. Table 4.3 shows the performance of using collective learning on CoNLL evaluation metrics. Table 4.4 shows performance of using collective learning on sentence-wide evaluation metrics.. We use ‘*’ to indicate the configuration has statistically significant improvement. with BIOSMILE use ‘**’ to indicate the configuration has statistically significant improvement with both BIOSMILE and the config. 2. configurations.. There are four different. The config. 3 incorporates BIOSMILE with collective learning;. 4 incorporate BIOSMILE + SRL patterns with collective learning.. the config.. Table 4.3 shows that the. config.3 and 4 applying collective learning outperform BIOSMILE and BIOSMILE + SRL pattern by F-score 1.91% and 1.65% on overall ARG respectively. 26. And, their improvements.
(36) on sentence-wide evaluation, in Table 4.4, are F-score 6.93% and 4.43% on overall ARG respectively, especially on ARGX with 7.54% and 6.93%.. These improvements indicate. that uses MLN to collectively learned SRL can improve both individual arguments and the sentence-wide argument.. Table 4.5 shows the costs of the time and memory.. Despite, the. performances of our system, the config. 4, is slightly lower than the config. 3, but costs are much lower than the config 3 on both time and memory. Table 4.3: The performances of SRL using collective learning. ARGX ARGM Overall ARG Configuration P. R. F. 立. 政 P 治R 大F. ΔF -. 82.96 62.65 71.39. ΔF -. P. R. F. 90.62 84.55 87.48. 2. 1 + Pattern. 89.69 87.70 88.68 +1.20* 74.20 68.33 71.14 -0.25. 3. 1 + CL. 89.88 88.88 89.37 +1.89* 80.00 68.15 73.60 +2.21* 87.13 82.48 84.74 +1.91**. 4. 2 + CL. 90.90 88.44 89.65 +2.17* 77.81 66.93 71.96 +0.57. 學. 1. BIOSMILE. ‧. ‧ 國. 88.59 77.78 82.83. ΔF. 85.10 81.71 83.37 +0.54*. Nat. y. 87.24 81.89 84.48 +1.65**. sit. n. er. io. Table 4.4: The performances of SRL on sentence-wide evaluation metrics. ARGX Overall ARG Configuration P R F ΔF P R F ΔF. al. Ch. e n g c h- i. iv n U51.78 51.41 51.60. 1. BIOSMILE. 71.83 70.41 71.11. 2. 1 + Pattern. 74.77 73.94 74.35 +3.24*. 3. 1 + CL. 78.75 78.54 78.65 +7.54** 58.58 58.57 58.57 +6.93**. 4. 2 + CL. 78.26 77.82 78.04 +6.93** 56.09 55.95 56.02 +4.42*. 27. -. 55.27 55.12 55.19 +3.59*. Table 4.5: The cost of time and memory. Configuration Train time Test time Memory 1. BIOSMILE 1 hr 10m 127.0ms 1130MB 2. 1 + Pattern 36 m 91.0ms 92MB 3. 1 + CL 2 hr 55m 143.0ms 1130MB 4. 2 + CL 1 hr 40m 103.0ms 92MB. -.
(37) 4.3 Related Work 4.3.1 Biomedical Semantic Role Labeling Corpus PASBio[21] is the first PAS standard used in the biomedical field, but it does not provide the SRL corpus. GREC[22] is an information extraction corpus focuses on gene regulation event.. However, GREC do not support the Treebank format SRL annotations[23].. BioProp is the only corpus that provides SRL annotations and annotates semantic role labels on the syntactic trees.. BioProp is created by [24].. BioProp selects 30 most frequently or. 政 治 大 Furthermore, following the style of PropBank[7], which annotates PAS on 立. important verbs appearing in the biomedical literatures, and defines the standard of the biomedical PAS.. Penn Treebank ( PTB ) [23], BioProp annotates their PAS on GENIA TreeBank ( GTB ) beta. ‧ 國. 學. version[25].. GTB contains a collection of 500 MEDLINE abstracts selected from the search. sit. y. Nat. contains the TreeBank that follows the style of Penn Treebank.. ‧. results with the following keywords: human, blood cells, and transcription factors and. io. al. er. 4.3.2 Biomedical Semantic Role Labeling System. v. n. Most semantic role labeling systems follow the pipeline method, which includes predicate. Ch. engchi. i n U. identification, argument identification and argument classification.. However, in recent years,. instead of using pipeline method, several researches have shown that using the collective learning method could outperform the pipeline method. collectively learned these stages on SRL.. However, we found that there seem to be no SRL. system using MLN in the biomedical field. improve SRL in biomedical field.. [20] uses Markov Logic to. [26] uses the domain adaption approaches to. [27] considers SRL as token-by-token labeling problem. and focuses on the SRL in the transport protein. BIOSMILE is the biomedical SRL system 28.
(38) focus on 30 frequently appearing or important verbs in biomedical literatures and trained on the BioProp, and it is based on Maximum Entropy ( ME ) Model.. 立. 政 治 大. ‧. ‧ 國. 學. n. er. io. sit. y. Nat. al. Ch. engchi. 29. i n U. v.
(39) CHAPTER 5 Conclusion We observe that some SRL ignore the complexity in classification and the dependencies between the semantic roles.. These systems usually take all constituents as candidate. 政 治 大. semantic roles and use a post-processing step to deal with their dependencies.. 立. In this paper,. to tackle both problems, we construct a biomedical SRL system that uses SRL patterns and a. ‧ 國. 學. Markov Logic Network ( MLN ) to collectively learned semantic roles. However, SRL patterns are difficult to be manually written, and we use automatically generated approaches,. ‧. to recognize the words boundaries and the candidates of semantic roles simultaneously.. Nat. y. Our. sit. system is trained on BioProp corpus. The experimental results show that using SRL patterns. n. al. er. io. can improve the performance by F-score 0.54% on overall ARG.. i n U. Furthermore, using. v. collective learning, which incorporated with linguistic constraints, can improve the result by F-score 1.65%.. Ch. engchi. We show that uses SRL patterns can improve the efficiency of training. model and predicate instances, and reduce the memory. can compete with current state-of-the-art approaches.. Also, we show that our approaches. The corpus used in our experiments is. a small biomedical SRL corpus that only uses one out of four of GENIA TreeBank corpus and also focuses on 30 verbs. future.. It is important to enable SRL to be trained on a large corpus in the. We consider that our approaches provide a possible solution to process large SRL. corpus.. 30.
(40) References. [2]. [3]. 立. 政 治 大. 學. ‧. [12]. Logic," in Proceedings of the 24th Annual Conference on Uncertainty in AI (UAI '08), ed, 2008, pp. 468-475. P. Domingos and M. Richardson, "Markov Logic: A Unifying Framework for. io. [7] [8] [9]. [10]. y. al. n. [6]. sit. [5]. Nat. [11]. K. B. Cohen and L. Hunter, "A critical review of PASBio's argument structures for biomedical verbs.," BMC Bioinformatics, vol. 7, 2006. S. Pradhan, K. Hacioglu, V. Krugler, W. Ward, J. H. Martin, and D. Jurafsky. (2005). Support Vector Learning for Semantic Argument Classification T. Cohn and P. Blunsom, "Semantic role labelling with tree conditional random fields," in In Proceedings of CoNLL-2005, ed, 2005, pp. 169-172. P. Kingsbury and M. Palmer, "From Treebank to PropBank," ed, 2002. X. Carreras and L. Marquez, "Introduction to the CoNLL-2005 Shared Task: Semantic Role Labeling," 2005. D. Gildea and M. Palmer, "The necessity of parsing for predicate argument recognition," in ACL '02: Proceedings of the 40th Annual Meeting on Association for Computational Linguistics, 2001, pp. 239-246. V. Punyakanok, D. Roth, W.-t. Yih, and D. Zimak, "Semantic role labeling via integer linear programming inference," in In Proceedings of COLING-04, ed, 2004, pp. 1346-1352. S. Riedel, "Improving the accuracy and Efficiency of MAP Inference for Markov. er. [4]. H.-J. Dai, Y.-C. Chang, R. Tzong-Han Tsai, and W.-L. Hsu, "New Challenges for Biological Text-Mining in the Next Decade," Journal of Computer Science and Technology, vol. 25, pp. 169-179, 2010. R. T.-H. Tsai, W.-C. Chou, Y.-S. Su, Y.-C. Lin, C.-L. Sung, H.-J. Dai, I. T.-H. Yeh, W. Ku, T.-Y. Sung, and W.-L. Hsu, "BIOSMILE: a semantic role labeling system for biomedical verbs using a maximum-entropy model with automatically generated template features," BMC Bioinformatics, vol. 8, p. 325, 2007. S. Pradhan, W. Ward, K. Hacioglu, J. Martin, and D. Jurafsky, "Shallow Semantic Parsing using Support Vector Machines," in Proceedings of the Human Language Technology Conference/North American chapter of the Association for Computational Linguistics annual meeting (HLT/NAACL-2004), Boston, MA, USA, 2004.. ‧ 國. [1]. Ch. engchi. 31. i n U. v.
(41) Statistical Relational Learning," in PROCEEDINGS OF THE ICML-2004 WORKSHOP ON STATISTICAL RELATIONAL LEARNING AND ITS CONNECTIONS TO OTHER FIELDS, 2004, pp. 49-54.. [14] [15] [16] [17] [18]. 立. 政 治 大. 學. ‧. [19]. M. Richardson and P. Domingos, "Markov logic networks," Machine Learning, vol. 62, pp. 107-136, 2006. K. Crammer and Y. Singer, "Ultraconservative online algorithms for multiclass problems," Journal of Machine Learning Research, vol. 3, pp. 951-991, 2003. S. Riedel, "Improving the accuracy and efficiency of map inference for markov logic," presented at the Proceedings of UAI 2008, 2008. D. Gildea and D. Jurafsky, "Automatic labeling of semantic roles," Comput. Linguist., vol. 28, pp. 245-288, 2002. N. Xue, "Calibrating features for semantic role labeling," in In Proceedings of EMNLP 2004, ed, 2004, pp. 88-94. M. Surdeanu, S. Harabagiu, J. Williams, and P. Aarseth, "Using predicate-argument structures for information extraction," presented at the Proceedings of the 41st Annual Meeting on Association for Computational Linguistics - Volume 1, Sapporo, Japan, 2003. R. Agrawal, T. Imieli\, \#324, ski, and A. Swami, "Mining association rules between. ‧ 國. [13]. sets of items in large databases," SIGMOD Rec., vol. 22, pp. 207-216, 1993.. Nat. [22]. [23] [24]. y. al. n. [21]. sit. io. S. Riedel and I. Meza-Ruiz, "Collective semantic role labelling with Markov logic," presented at the Proceedings of the Twelfth Conference on Computational Natural Language Learning, Manchester, United Kingdom, 2008. T. Wattarujeekrit, P. Shah, and N. Collier, "PASBio: predicate-argument structures for event extraction in molecular biology," BMC Bioinformatics, vol. 5, p. 155, 2004. P. Thompson, S. Iqbal, J. McNaught, and S. Ananiadou, "Construction of an annotated corpus to support biomedical information extraction," BMC Bioinformatics, vol. 10, p. 349, 2009. A. Bies, "Bracketing Guidelines for Treebank II Style Penn Treebank Project," ed, 1995. W.-C. Chou, R. T.-H. Tsai, Y.-S. Su, W. Ku, T.-Y. Sung, and W.-L. Hsu, "A semi-automatic method for annotating a biomedical proposition bank," presented at the Proceedings of the Workshop on Frontiers in Linguistically Annotated Corpora 2006, Sydney, Australia, 2006.. er. [20]. Ch. engchi. i n U. v. [25]. J. D. Kim, T. Ohta, Y. Tateisi, and J. Tsujii, "GENIA corpus -- a semantically annotated corpus for bio-textmining," Bioinformatics, vol. 19, pp. i180-i182, 2003.. [26]. D. Dahlmeier and H. T. Ng, "Domain Adaptation for Semantic Role Labeling in the Biomedical Domain," Bioinformatics (Oxford, England), 2010. 32.
(42) S. Bethard, Z. Lu, J. Martin, and L. Hunter, "Semantic Role Labeling for Protein Transport Predicates," BMC Bioinformatics, vol. 9, p. 277, 2008.. 立. 政 治 大. 學 ‧. ‧ 國 io. sit. y. Nat. n. al. er. [27]. Ch. engchi. 33. i n U. v.
(43)
數據

相關文件
了⼀一個方案,用以尋找滿足 Calabi 方程的空 間,這些空間現在通稱為 Calabi-Yau 空間。.
• ‘ content teachers need to support support the learning of those parts of language knowledge that students are missing and that may be preventing them mastering the
refined generic skills, values education, information literacy, Language across the Curriculum (
volume suppressed mass: (TeV) 2 /M P ∼ 10 −4 eV → mm range can be experimentally tested for any number of extra dimensions - Light U(1) gauge bosons: no derivative couplings. =>
• Formation of massive primordial stars as origin of objects in the early universe. • Supernova explosions might be visible to the most
• Flux ratios and gravitational imaging can probe the subhalo mass function down to 1e7 solar masses. and thus help rule out (or
This kind of algorithm has also been a powerful tool for solving many other optimization problems, including symmetric cone complementarity problems [15, 16, 20–22], symmetric
(Another example of close harmony is the four-bar unaccompanied vocal introduction to “Paperback Writer”, a somewhat later Beatles song.) Overall, Lennon’s and McCartney’s