Research Article
Lead Screening for Chronic Obstructive Pulmonary Disease of
IKK2 Inhibited by Traditional Chinese Medicine
Yung-An Tsou,
1,2Hung-Jin Huang,
3Wesley Wen-Yang Lin,
4and Calvin Yu-Chian Chen
2,51Otolaryngology Head and Neck Surgery, China Medical University Hospital, Taichung 40402, Taiwan 2School of Medicine, College of Medicine, China Medical University, Taichung 40402, Taiwan
3Department of Chinese Pharmaceutical Sciences and Chinese Medicine Resources, College of Pharmacy, China Medical University,
Taichung 40402, Taiwan
4Department of Biological Science and Technology, National Chiao Tung University, Hsinchu 30010, Taiwan 5Department of Biomedical Informatics, Asia University, Taichung 41354, Taiwan
Correspondence should be addressed to Calvin Yu-Chian Chen; ycc929@MIT.edu
Received 17 January 2014; Revised 8 February 2014; Accepted 8 February 2014; Published 2 June 2014 Academic Editor: Fuu-Jen Tsai
Copyright © 2014 Yung-An Tsou et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Chronic obstructive pulmonary disease (COPD) is a chronic obstructive lung disease and is frequently found in well-developed countries due to the issue of aging populations. Not all forms of medical treatment are unable to return a patient's limited pulmonary function back to normal and eventually they could require a lung transplant. At this time, COPD is the leading cause of death in the world. Studies surveying I-kappa-B-kinase beta (IKK2) are very relevant to the occurrence and deterioration of the condition COPD. The sinapic acid-4-O-sulfate, kaempferol, and alpha-terpineol were found to be IKK2 inhibitors and helped prevent COPD occurrence and worsening according to a screening of the traditional Chinese medicine (TCM) database. The protein-ligand interaction of these three compounds with regard to IKK2 was also done by molecular dynamics. The docking poses, hydrogen bond variation, and hydrophobic interactions found Asp103 and Lys106 are crucial to IKK2 binding areas for IKK2 inhibition. Finally, we found the three compounds that have an equally strong effect in terms of IKK2 binding proven by the TCM database and perhaps these may be an alternative treatment for COPD in the future.
1. Introduction
COPD, a chronic obstructive pulmonary disease, can lead to the restriction of lung function [1,2]. The current treatment options for COPD are very limited and the side effect of treatment frequently noted is Cushing syndrome caused by long term steroid use [3]. Many COPD patients finally need lung transplants and the survival outcome is still poor even when patients undergo lung transplants [4]. Even with improvement in regard to pharmacy and drug invention techniques, the occurrence of COPD and mortality related to COPD continues to rise [5]. Clearly, efforts to prevent smoking, reduce air-pollution, and control pneumonia could be the appropriate prevention methods to limit deterioration in cases of COPD. However, there are no other useful ways to
attempt to cure the COPD; thus it remains the leading cause of death throughout the world [2,6]. Therefore, prevention of the occurrence of COPD is the important issue to address, by not only the above mentioned methods but also the inhibition of I-kappa-B-kinase beta (IKK2) which is linked to COPD occurrence [7–10].
IKK2 activation is related to many inflammatory dis-eases, severe immune compromise diseases [11], severe skin infection [12,13], chronic intestinal infection [14], synovial inflammation [15], arthritis [16], pancreatitis, and diabetes mellitus [17]. There have been other reports that highlighted IKK2 activation as leading to chronic airway diseases such as Lipopolysaccharides (LPS) related airway infections, asthma, emphysema, and chronic bronchitis [7,18]. The very impor-tant subunit of I𝜅B kinase is IKK2 that has an enzyme Volume 2014, Article ID 465025, 16 pages
Table 1: Scoring functions of top high ranking candidates from docking results.
Name Dock Score -PLP1 -PLP2 -PMF
Sinapic acid-4-O-sulfate 189.61 62 60.93 48.9 Kaempferol 174.852 54 47.17 44.31 𝛼-Terpineol 157.438 39 33.49 25.96 Geranial 150.458 42 43.71 22.91 3-(2-Carboxyphenyl)-4(3H)-quinazolinone 147.919 54 46.9 33.24 Arctigenin 145.065 71 72.46 85.33 Notoginsenoside G 143.481 54 49.1 38.14 ∗Staurosporine 115.865 141 129.69 117.56 ∗Control. O O O S OH O O− O− −O (a) O NH −O +H 3N (b) O N H N O− H2+ (c) N O N HO HN O O (d)
Figure 1: The scaffold of top three TCM compounds and control: (a) sinapic acid-4-O-sulfate (b), kaempferol (c), alpha-terpineol (d), and staurosporine.
function that is involved in activating the NF-kB (nuclear transcription factor kappa-B) and causing the subsequent inflammatory condition IKK2 could be phosphorated by a protein called the I𝜅B (I𝜅B𝛼), which could inhibit the NF-𝜅B by direct binding. Ubiquitination process is then started after I𝜅B phosphorylation, and then I𝜅B is degraded and set free of NF-𝜅B into the nucleus and causes subsequent immune and inflammation responses [16,17,19].
Utilizing a computational simulation technique could efficiently help identify suitable drugs for IKK2 inhibition through the use of computer-aided drug design (CADD)
which can help to structure the best drug candidates and pre-dict biological activity. CADD is a very efficient way to treat any specific disease with appropriate drugs targeting [20]. The novel targeting factors for drug design should be based on some related studies [21–27] and also include risk factor anal-ysis [28–32]. This method is also advantageous in terms of economic efficiency [33]. The drug design by its structure and ligand-based binding survey are two essential components of CADD [34–38]. Then, the appropriate drug candidates are screened by molecular simulation that are structure-based and confirm the molecular dynamics in CADD [39].
Cys99 Val152 Val29 Ile165 Gly22 Leu21 Glu149 Asp103 Asp106 Gly102 Tyr98 Sigma pi (a) Cys99 Val152 Tyr98 Ile165 Ala42 Val49 Leu21 Gly22 Thr23 Asp103 Arg105 Glu149 Gly102 (b) Asp103 Gly22 Arg105 Leu21Leu149 Thr23 Lys106 Ile165 Val152 Cys99 Gly102 Tyr98 (c) Lys147 Asn105 Thr23 Gly22 Glu149 Asp166 Lys44 Leu94 Leu61 Val29 Met96 Val74 Glu97Ala42 Asp103 Leu21 Gly102 Val152 Cys99 Tyr98 Ile165 Sigma pi Sigma pi (d)
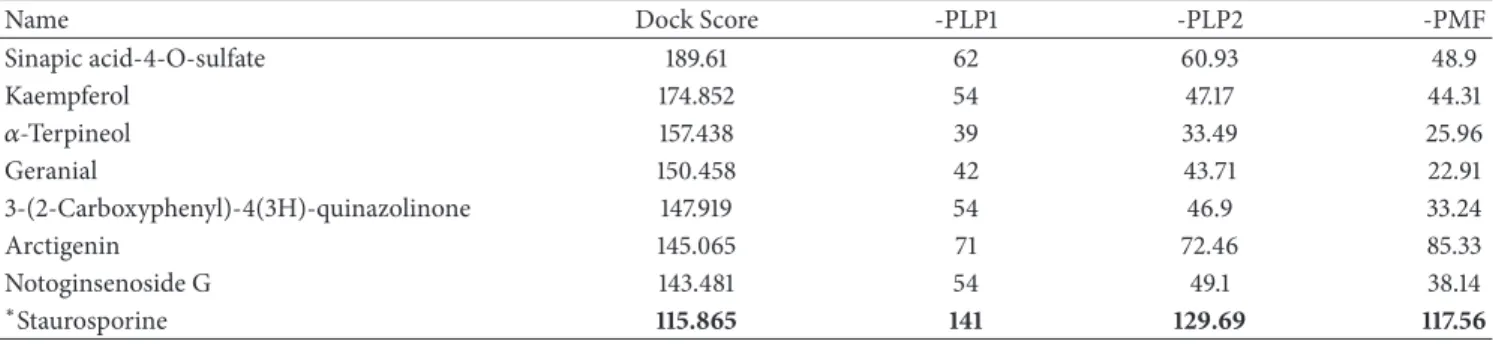
Figure 2: The ligands and poses of docking site for IKK2 and its docking site crystal structure: (a) sinapic acid-4-O-sulfate, (b) kaempferol, (c) alpha-terpineol, and (d) staurosporine.
Table 2: H-bond occupancy among all MD simulations.
Compound name Atoms of H-bonds Occupancy Compound name Atoms of H-bonds Occupancy
Sinapic acid-4-O-sulfate O14:LIG—NZ:LYS106 10.76% Kaempferol HZ3:LYS106—O14:LIG 18.33% HZ3:LYS106—O15:LIG 42.23% HZ3:LYS106—O15:LIG 59.76% O7:LIG—N:GLY22 0.40% HN:ASP103—O15:LIG 48.21% HN:GLY22—O17:LIG 0.40% HH:TYR98—O14:LIG 59.36% O18:LIG—N:GLY22 5.58% H27:LIG—O:LEU21 23.51% HN:GLY22—O20:LIG 12.35% H27:LIG—OD1:ASP103 74.90% — — H27:LIG—OD2:ASP103 47.01% — — H27:LIG—OE2:GLU149 9.16% — — H27:LIG—O:GLU149 14.34% alpha-Terpineol HG1:THR23—O13:LIG 29.48% Staurosporine O25:LIG—N:THR23 0.00% HG1:THR23—O14:LIG 51.00% HN:THR23—O1:LIG 59.36% HH22:ARG20—O13:LIG 63.35% O:GLU97—O6:LIG 0.00% HH22:ARG20—O14:LIG 54.58% — — HH12:ARG20—O13:LIG 55.38% — — HH12:ARG20—O14:LIG 47.01% — — HE:ARG20—O13:LIG 2.79% — — HE:ARG20—O14:LIG 4.38% — — H30:LIG—OD1:ASP103 3.98% — — H30:LIG—OD2:ASP103 3.19% — — H22:LIG—OE1:GLU149 15.54% — — H22:LIG—OE2:GLU149 16.33% — — ∗LIG: ligand.
C4 C5 C6 C9 C10 C13 C1 C2 C3 O7 C8 O11 C12 O14 O15 S16 O17 O18 O19 O20 Asp103 Val29 Lys106 Leu21 Val152 Cys99 Gly102 Ile165 Glu149 (a) 2.70 C1 C2 C3 C4 C5 C6 N7 C8 C9 N10 C11 C12 C13 O14 O15 N CA CB CG OD1 OD2 C O Asp103 Leu21 Val152 Gly22 (b) C1 C2 C3 C4 C5 C6 N7 C8 C9 C10 C11 C12 O13 O14 N16 C17 Asp103 Leu21 Val152 Gly22 Gly102 Glu149 Ile165
Nonligand residues involved
Corresponding atoms involved in hydrophobic contact(s)
in hydrophobic contact(s) Ligand bond
Nonligand bond Hydrogen bond and its length
(c)
Nonligand residues involved
Corresponding atoms involved in hydrophobic contact(s)
in hydrophobic contact(s) Ligand bond
Nonligand bond Hydrogen bond and its length
2.70 2.86 2.82 N10 C11 C12 N13 C16 C17 C18 C19 C20 C21 C22 C23 C26 C29 C30 C31 C32 C33 C34 C35 O1 C2O3 C4 C5 O6 C7 O8 C9 C14 C15 C24 O25 C27 N28 N CA CB CG CD OE1 OE2 C O Glu149 N CA CB SG C O Cys99 Ile165 Leu21 N CA CB CG CD OE1 OE2 C O Glu97 Asp166 Gly22 Val29 Tyr98 Ala42 Gly102 Val152 Thr23 Met96 (d)
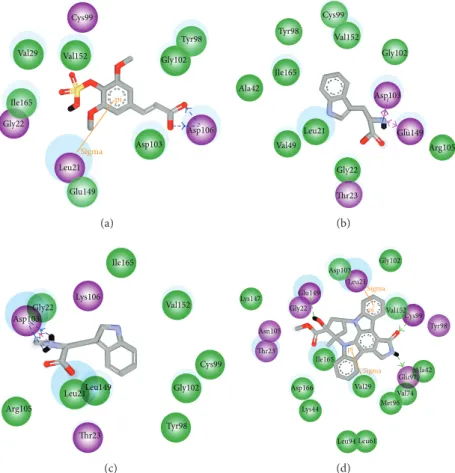
Figure 3: Protein-ligand interactions by Ligplot plus: (a) sinapic acid-4-O-sulfate, (b) kaempferol, (c) alpha-terpineol, and (d) staurosporine. The high frequency hydrophobic interactions, ligands’ interactions, were showed by deep red color spots.
0.5 1 0 100 200 300 400 500 600 700 Dis o rd er dis p osi tio n Residue index Disorder disposition Fe at u re s Residuex21–165
active binding domain
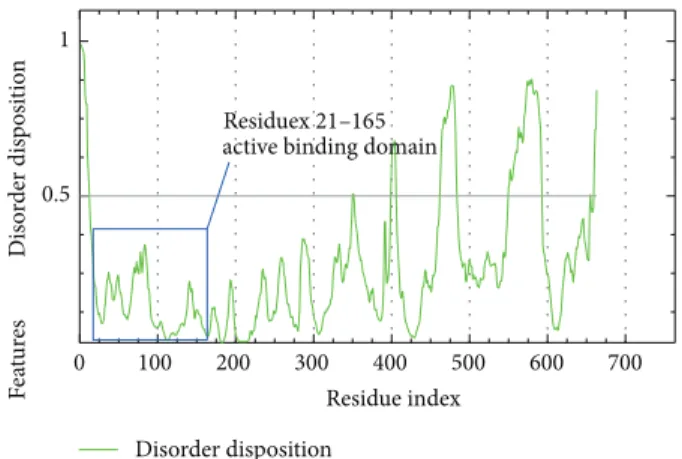
Figure 4: PONDR-FIT prediction of IKK2 active binding domain, value of disorder disposition below 0.5 indicated order residues.
Traditional Chinese medicine (TCM) has been used in China, Taiwan, Korea, and Japan for thousands of years. The largest traditional Chinese medicine database thus far is the TCM Database@Taiwan (http://tcm.cmu.edu.tw/) [39] which contains 61,000 compounds of Chinese herbal medicine with their 2D and 3D chemical structures, molecular information, and bioactivity. Novel lead compounds have been found for the treatment of cancer [40,41], controlling pain [42], and virus detoxification [33,34,43] by TCM Database@Taiwan since 2011. The TCM Database@Taiwan is an important tool that will help to determine TCM drug design [21, 44–51] to help overcome related clinical difficulties and it could be accessed by the website portal [33] or through the cloud computing platform [52].
Based on recent study, a possible lead compound for COPD treatment has been identified by the TCM
0 1000 2000 3000 4000 5000 Sinapic acid-4-O-sulfate Kaempferol alpha-Terpineol Staurosporine Time (ps) 0.35 0.30 0.25 0.20 0.15 0.10 P ro tein RMS D (nm) (a) Sinapic acid-4-O-sulfate Kaempferol alpha-Terpineol Staurosporine 0 1000 2000 3000 4000 5000 64 62 60 58 56 54 52 P ro tein SAS (nm-S 2-N) Time (ps) (b) Sinapic acid-4-O-sulfate Kaempferol alpha-Terpineol Staurosporine 0 1000 2000 3000 4000 5000 2.06 2.04 2.02 2.00 1.98 1.96 P ro tein GYRA T E (nm) Time (ps) (c)
Figure 5: Trajectories of (a) complex RMSD, (b) SASA, and (c) gyrate.
Database@Taiwan in this study. The docking screening of selected COPD ligands is done by utilizing computational techniques and confirming the molecular dynamics (MD) for protein-ligand interactions affected the most regarding IKK2 inhibitions in COPD.
2. Materials and Methods
2.1. Data Set. Molecular simulations were performed by
Accelrys Discovery Studio 2.5 (DS 2.5) system. There were 61,000 TCM compounds downloaded from TCM database
(http://tcm.cmu.edu.tw/) analyzed in this study. The IKK2
(PDB ID: 4KIK) [53] crystal structure from the RCSB Protein Data Bank was used for research, and the staurosporine was used as a control.
2.2. Molecular Dockings. LigandFit [54] was used to dock
SC-514 staurosporine and TCM compounds to IKK2 in the CHARMm force field for docking simulation [55]. LigandFit
is a program set on Discovery Studio 2.5 (DS 2.5) for docking receptor-rigid algorithm. The IKK2 docking site was surveyed and the top 3 compounds according to the docking score were obtained and Ligplot plus program was used for their hydrophobic interactions analysis [56,57]. After that, we evaluated the protein-ligand interaction and drug efficacy by comparing the disorder region and the docking site.
2.3. Detection of the Disordered Protein. Disorder region was
predicted according to protein structure and docking site by the Database of Protein Disorder (DisProt,http://www
.disprot.org/). Thus, we could prevent disorder effects on drug
design and determine the docking site and the efficacy of the drug more precisely [54,58].
2.4. Simulation of Molecular Dynamics. The surveyed
lig-ands prepared for further MD simulation were supported by SwissParam (http://swissparam.ch/) [55] on the basis of reference force field [56] counted by GROMACS 4.5.5 [57].
RMS
F (nm)
Sinapic acid-4-O-sulfate 0.4 0.3 0.2 0.1 0.0 0 50 100 150 200 250 300 Residue (a) Kaempferol RMS F (nm) 0.3 0.2 0.1 0.0 0 50 100 150 200 250 300 Residue (b) alpha-Terpineol Residue RMS F (nm) 0.4 0.3 0.2 0.1 0.0 0 50 100 150 200 250 300 (c) Staurosporine Residue RMS F (nm) 0.3 0.2 0.1 0.0 0 50 100 150 200 250 300 (d)
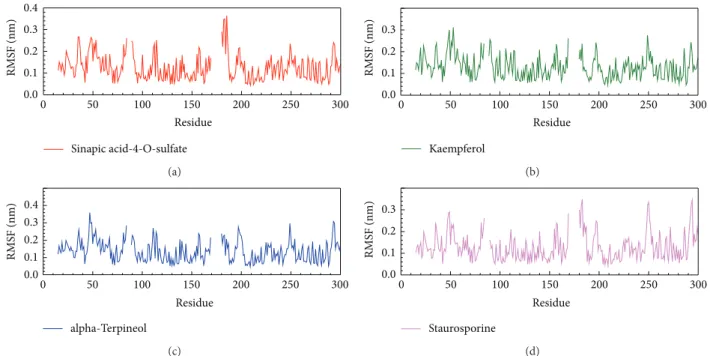
Figure 6: The variation of staurosporine and IKK2 complex in RMSF MD simulation: (a) sinapic acid-4-O-sulfate, (b) kaempferol, (c) alpha-terpineol, and (d) staurosporine.
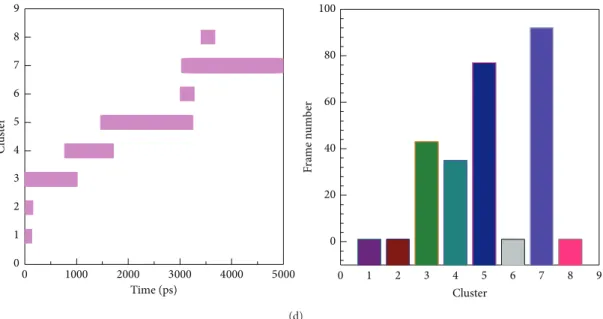
Table 3: Time of middle frames in each cluster.
Cluster Time of middle frame (ps)
Sinapic acid-4-O-sulfate Kaempferol alpha-Terpineol Staurosporine
1 200 0 0 0 2 380 20 780 20 3 1900 140 2840 520 4 — 1320 4940 1280 5 — 3620 — 2460 6 — 4920 — 3140 7 — — — 3960 8 — — — 3540
The docked ligands and IKK2 protein complex were set into the simulation box in the buffer. The minimum distance was set at 1.2 ˚A from the complex in the cubic box and then solvated in TIP3P water circumstances in which the complex charge was neutralized by adding sodium and chlo-ride. The steepest descent method was used for minimizing complex for 5000 steps. The structure after minimization was finally used for MD simulation. The particle-mesh Ewald (PME) method was then used for calculating the electro-static interactions between the ligands and IKK2 complex [59]. Each step was set for 2 fs and 2,500,000 steps were performed by the PME method. The Berendsen weak thermal coupling method was used for equilibration under constant temperature (NVT ensemble) for 100 ps was performed. The total time was set to 5000 ps for MD simulation process. Then the protocol in Gromacs was finally applied for MD trajectories, RMSD, and energy variations of complex were surveyed.
3. Results and Discussions
3.1. Docking Results for Molecules. The top 3 TCM
com-pounds were selected (Table 1) by molecular docking accord-ing to their dockaccord-ing scores. These TCM compounds are sinapic acid-4-O-sulfate, kaempferol, and alpha-terpineol belonging to the TCM herbs, the bark of Magnolia officinalis,
Bupleurum chinense, and Bursaphelenchus xylophilus. The
first compound, sinapic acid-4-O-sulfate, has an antidepres-sant like effect [60, 61] and the herb, the bark of Magnolia
officinalis, has potent anti-inflammation effects to prevent
further tissue inflammation [62, 63]. The 2nd compound, the herb Bupleurum chinense, can be hepatoprotective, anti-inflammatory, analgesic, and antipyretic and also prevent acute lung injury and just be a good treatment material for COPD [64]. The 3rd compound alpha-terpineol and herb Bursaphelenchus xylophilus have an antimicrobial effect and in particular prevent infections that originate from
−822000 −824000 −826000 −828000 −830000 −832000 Time (ns) T o ta l ener g y (kJ/mo l) 0 1000 2000 3000 4000 5000
Sinapic acid-4-O-sulfate
(a) −822000 −824000 −826000 −828000 −830000 T o ta l ener g y (kJ/mo l) Time (ns) 0 1000 2000 3000 4000 5000 Kaempferol (b) −822000 −824000 −826000 −828000 −830000 T o ta l ener g y (kJ/mo l) Time (ns) 0 1000 2000 3000 4000 5000 alpha-Terpineol (c) −822000 −824000 −826000 −828000 −830000 T o ta l ener g y (kJ/mo l) Time (ns) 0 1000 2000 3000 4000 5000 Staurosporine (d)
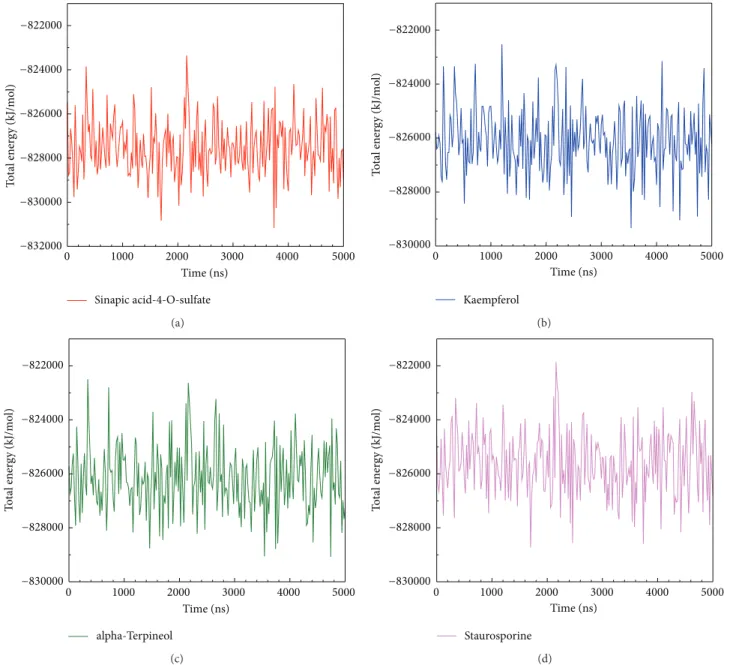
Figure 7: The MD simulation showed the binding energy variation of top three compounds and control to IKK2 complex: (a) sinapic acid-4-O-sulfate, (b) kaempferol, (c) alpha-terpineol, and (d) staurosporine.
periodontopathic and cariogenic bacteria [65]. Most of these compounds can prevent humans from further infection and inflammation and could be protectors for lung injury.
The candidate compounds and control structure were selected after screening the TCM database (Figure 1). After that, the amino acid neighbors by ligand docking site were displayed inFigure 2. We found Asp103, Leu21, and Cys99 are amino acids that could interact with control and selected compound ligands. We considered these three amino acids as playing a crucial role with regard to IKK2 target func-tion.
The Ligplot plus [66] applied for hydrophobic interaction ligand-compound complex survey (Figure 3). The amino
acids Glu149, Val29, Ile165, Val152, Gly102, and Gly22 pre-sented deep red color showing high frequency protein-ligand interactions by hydrogen bond or hydrophobic interactions. These amino acids are very crucial for reference and we selected compounds which were proven by hydrophobic interaction analyses that have an effect on IKK2.
3.2. The Results of Disorder Proteins Detection. There were
important amino acids nearby the docking site for IKK2 docking which include the Asp103, Leu21, Cys99, Glu149, Val29, Ile165, Val152, Gly102, and Gly22. They were consid-ered as active ATP binding sites in IKK2. Staurosporine can
Resid u e index 250 200 150 100 50 Residue index 50 100 150 200 250 Distance 0 1.5 (a) Resid u e index 250 200 150 100 50 Residue index 50 100 150 200 250 Distance 0 1.5 (b) Resid u e index 250 200 150 100 50 Residue index 50 100 150 200 250 Distance 0 1.5 (c) Resid u e index 250 200 150 100 50 Residue index 50 100 150 200 250 Distance 0 1.5 (d)
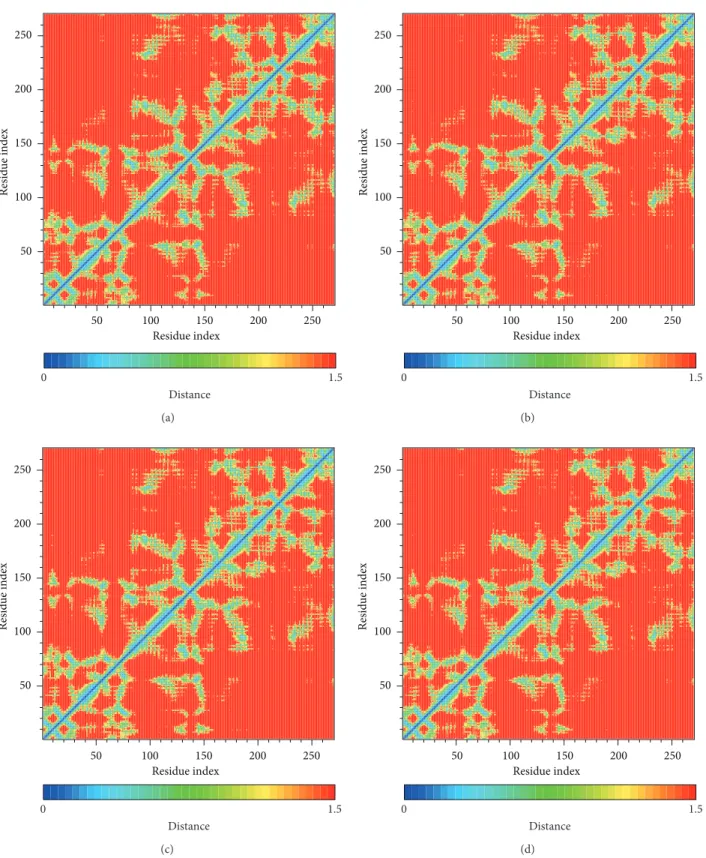
Figure 8: The residue index of bindings distances change: (a) sinapic acid-4-O-sulfate, (b) kaempferol, (c) alpha-terpineol, and (d) staurosporine.
250 200 150 100 50 Resid ue 0 1000 2000 3000 4000 5000 Time (ps) 0 1000 2000 3000 4000 5000 Time (ps) 60 50 40 30 20 10 0 St ru ct ur e f ea tur es (%) 𝛽-Sheet𝛼-Helix Turn Others (a) 250 200 150 100 50 Resid ue 0 1000 2000 3000 4000 5000 Time (ps) 0 1000 2000 3000 4000 5000 Time (ps) 60 50 40 30 20 10 0 St ru ct ur e f ea tur es (%) 𝛽-Sheet𝛼-Helix Turn Others (b)
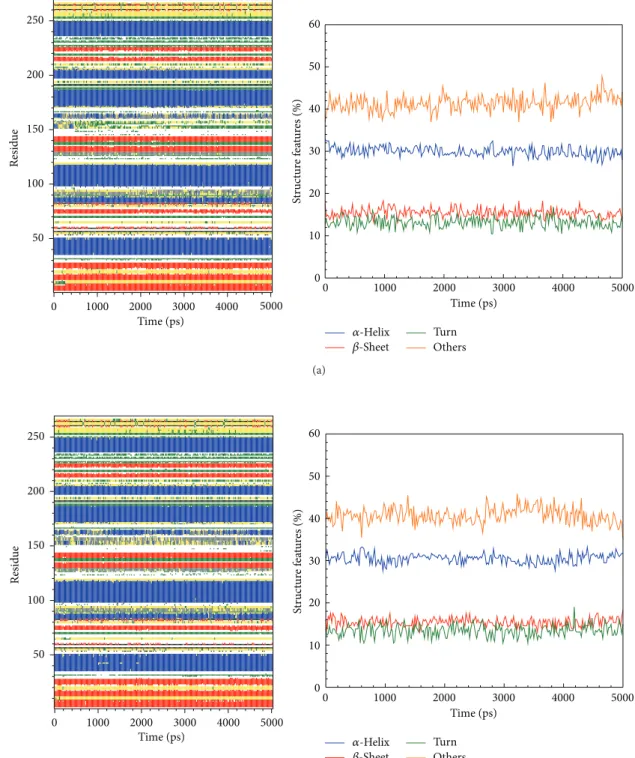
Figure 9: The variation of secondary structural changes during binding process showed by MD simulation of top candidates to IKK2: (a) sinapic acid-4-O-sulfate and (b) kaempferol.
inhibit IKK2 by these regions through the binding of targets and as a reference to other compounds. Disorder prediction shows that the binding residues 21 to 165 are all located in ordered region below 0.5 disorder disposition as showed in
Figure 4.
3.3. Simulation for Molecular Dynamics. The trajectory of
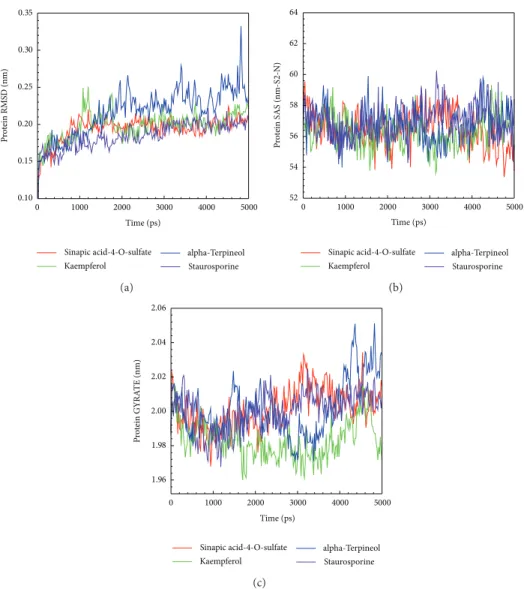
protein-ligand complexes were calculated during MD sim-ulation (Figure 5). All of the three top compounds, sinapic acid-4-O-sulfate kaempferol, and alpha-terpineol, with low variation showed in the complex RMSD for the IKK2
(c) (d) 0 1000 2000 3000 4000 5000 Time (ps) 250 200 150 100 50 Resid ue 0 1000 2000 3000 4000 5000 Time (ps) 60 50 40 30 20 10 0 St ru ct ur e f ea tur es (%) 𝛽-Sheet 𝛼-Helix Turn Others 0 1000 2000 3000 4000 5000 Time (ps) 250 200 150 100 50 Resid ue 0 1000 2000 3000 4000 5000 Time (ps) 60 50 40 30 20 10 0 St ru ct ur e f ea tur es (%) 𝛽-Sheet𝛼-Helix Turn Others
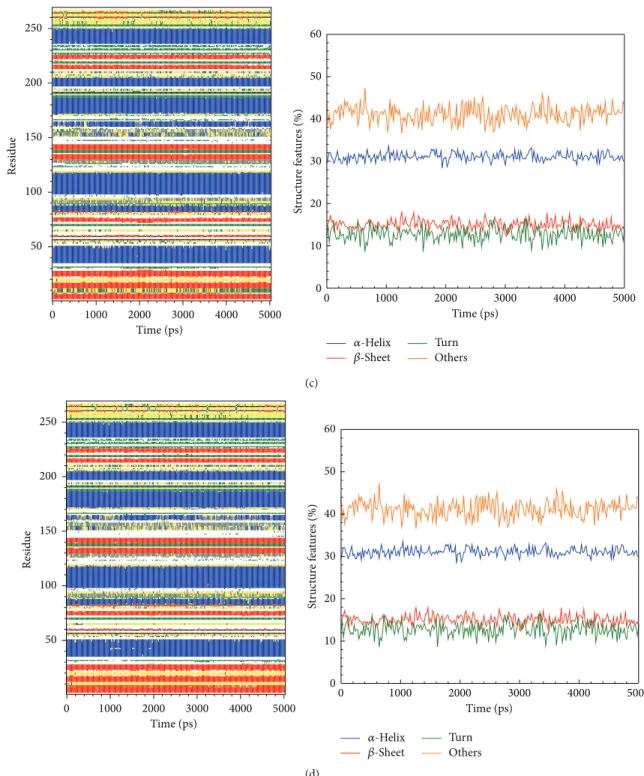
Figure 10: The variation of secondary structural changes during binding process showed by MD simulation of top candidate and control to IKK2: (c) alpha-terpineol and (d) staurosporine.
phosphorylation interaction. The stable distance tended to be 0.25 nm of the complex RMSD showing a stable and balanced complex interaction. SASA analysis showing that the solvent area revealed a stable in IKK2 after the three compounds bindings in Figure 5(b). Gyration displayed a stable fluctuation between 1.96 and 2.04 nm during the IKK2
bindings to each of the three compounds during the MD simulation.
In stability analysis of each residue on the binding region over MD simulation, the major binding regions are located on 21 to 149 residues (Figure 6), and we found that the RMSF values are not substantial fluctuations and are similar to each
Time (ps) 5000 4000 3000 2000 1000 0 Cl u st er 4 3 2 1 0 200 240 150 100 50 0 0 1 2 3 4 F ra m e n u m b er Cluster (a) Time (ps) 5000 4000 3000 2000 1000 0 Cl u st er 5 6 7 3 4 2 1 0 120 140 60 80 100 40 20 0 F ra m e n u m b er 0 1 2 3 4 5 6 7 Cluster (b) Time (ps) 5000 4000 3000 2000 1000 0 Cl u st er 5 3 4 2 1 0 140 100 50 0 0 1 2 3 4 5 F ra m e n u m b er Cluster (c) Figure 11: Continued.
Time (ps) 5000 4000 3000 2000 1000 0 Cl u st er 5 6 9 8 7 3 4 2 1 0 60 80 100 40 20 0 F ra m e n u m b er 0 1 2 3 4 5 6 7 8 9 Cluster (d)
Figure 11: Further clustering analysis of the hit component for further IKK2 binding MD survey.
other, exhibiting stable conformation during MD simulation. The trajectories of total energy by MD measurement are presented inFigure 7which shows that the three top com-pounds and the staurosporine binding to form IKK2 complex have no significant difference in total energy compared to each other. The ranges of total energy were between−822000 and−820000 kJ/mol and stabilized to −868000 kJ/mol. This presented that the complex is still in a stable still interaction even after a 5000 ps as showed inFigure 7.
We calculated distance for pair of each residue during all simulation time; there is no significant difference between all protein-ligand complexes, indicating that complexes remain stable during the simulation time (Figure 8). In DSSP analysis (Figures9and10), the MD simulation showed IKK2 as having three different binding phases. The IKK2 is a transmembrane protein with no change in the structure (helix becoming a loop or others) of the three compounds and staurosporine when binding to IKK2. In Figures9and10, we found these three compounds were all potent candidates for the treatment of COPD patients and all the compounds and control did not have their structure composition broken and had a stable complex interaction.
In addition, the distance of H-bond affecting the occu-pancy of H-bond was also calculated during MD (Table 2). We found the important amino acids Asp103 and Lys106 having high occupancy H-bond formation between IKK2 phosphorylation sides and sinapic acid-4-O-sulfate and kaempferol. This may explain why these amino acids are important as they act on IKK2 phosphorylation and cause inhibition of NF-KB related inflammation.
After that, we clustered all MD structures using linkage algorithm to identify represented conformation for interac-tion analysis. All MD frames of four complexes with docked ligand were clustered to different groups (Figure 11), and the
middle structures of each group are represented inTable 3. We selected the largest group in latest MD time as repre-sented structure for snapshots study. In Figure 12(a), snap-shots of sinapic acid-4-O-sulfate forms H-bond interactions with Gly22 and Lys106. Figure 12(b) presented kaempferol forms H-bond interaction with Asp 103, Lys106, and Glu149.
Figure 12(c)showed alpha-terpineol forms H-bond
interac-tion with Arg20. Figure 12(d) revealed that staurosporine generates H-bond interaction with Thr23. Comparing with initial docking pose of sinapic acid-4-O-sulfate revealed H-bond interaction with the same Lys106 showed inFigure 2(a). Therefore, there is the same residue after MD simulation for sinapic acid-4-O-sulfate.
For docking pose of kaempferol in Figure 2(b), Glu149 and Asp103 were the initial binding residues. However, only Asp103 keeps the same stable residue in the H-bond interaction after MD simulation. It is worthy to notice that the initial binding poses of alpha-terpineol and staurosporine are different after MD simulation showed in Figures 3(c)
and 3(d). The results suggest sinapic acid-4-O-sulfate and kaempferol are more potent compounds candidate for IKK2 interaction. Besides, we further analyzed the ligand pathway during MD simulation. The prediction results were presented
inFigure 13; we found that (a) sinapic acid-4-O-sulfate, (b)
kaempferol, and (c) alpha-terpineol all have ligand channels. However, there was no ligand path for staurosporine; it is possible that the staurosporine is too compact to generate a ligand channel. We considered that these three compounds could really interact with IKK2 and consequently affect the related function of IKK2 during the binding process.
By TCM targeting IKK2 when drug screening, we found the three compounds from Chinese medicine to treat the COPD and believe that this may help clinicians select potent medicine to prevent patients from having COPD in the future
Gly22 Lys106 1900 ps (a) Tyr98 Leu21 Glu149 Asp103 Lys106 4920 ps (b) Thr23 Arg20 Glu149 Asp103 4940 ps (c) Thr23 3960 ps (d)
Figure 12: Snapshots of initial conformation and represented structures: (a) sinapic acid-4-O-sulfate, (b) kaempferol, (c) alpha-terpineol, and (d) staurosporine.
or to assist in the area of disease control for COPD. This identification method can also be useful for many other infectious or inflammatory diseases in terms of selecting the proper drugs for difficult treating diseases.
4. Conclusion
Based on the above discussion, we identified the top 3 TCM compounds, sinapic acid-4-O-sulfate, kaempferol, and alpha-terpineol, which can have an effect on IKK2 inhibition and prevent exacerbation and disease progression with regards to COPD. Asp103, Leu21, Cys99, Glu149, Cal29, Val152, Gly22,
and Gly102 108 present their crucial effect on IKK2 inhibition through H-bond formation and hydrophobic interaction. The Asp103 and Lys106 are very important residues in IKK2 bind-ing. These top three compounds can bind to the IKK2 ATP binding site and cause IKK2 inhibition by phosphorylation and may be used in future considerations in the development of novel therapies for COPD.
Conflict of Interests
The authors reaffirm that there is no conflict of interests to declare.
(a) (b)
(c) (d)
Figure 13: Prediction of ligand channels of the hit component during MD simulation.
Authors’ Contribution
Yung-An Tsou, Hung-Jin Huang, and Wesley Wen-Yang Lin contributed equally to this paper.
Acknowledgments
The research was supported by grants from the National Sci-ence Council of Taiwan (2325-B039-001, NSC102-2221-E-468-027, and NSC101-2314-B-039-013-MY3), from Asia University (ASIA100-CMU-2, ASIA101-CMU-2, and 102-ASIA-07), and from China Medical University Hospital (DMR-102-003, DMR-103-025, DMR-103-058, DMR-103-001, and DMR-103-096). This study is also supported in part by Taiwan Department of Health Clinical Trial and Research Center of Excellence (DOH102-TD-B-111-004) and Taiwan Department of Health Cancer Research Center of Excellence
(MOHW103-TD-B-111-03), and CMU under the Aim for Top University Plan of the Ministry of Education, Taiwan.
References
[1] G. E. Taffet, J. F. Donohue, and P. R. Altman, “Considerations for managing chronic obstructive pulmonary disease in the elderly,” Clinical Interventions in Aging, vol. 9, pp. 23–30, 2014. [2] J. Vestbo, S. S. Hurd, A. G. Agust´ı et al., “Global strategy for the
diagnosis, management, and prevention of chronic obstructive pulmonary disease GOLD executive summary,” The American
Journal of Respiratory and Critical Care Medicine, vol. 187, no. 4,
pp. 347–365, 2013.
[3] F. J. Martinez, J. F. Donohue, and S. I. Rennard, “The future of chronic obstructive pulmonary disease treatment—difficulties of and barriers to drug development,” The Lancet, vol. 378, no. 9795, pp. 1027–1037, 2011.
[4] S. Bustacchini, C. Chiatti, G. Furneri, F. Lattanzio, and L. G. Mantovani, “The economic burden of chronic obstructive pul-monary disease in the elderly: results from a systematic review of the literature,” Current Opinion in Pulmonary Medicine, vol. 17, no. 1, pp. S35–S41, 2011.
[5] M. Najafzadeh, C. A. Marra, L. D. Lynd et al., “Future impact of various interventions on the burden of COPD in Canada: a dynamic population model,” PLoS ONE, vol. 7, no. 10, Article ID e46746, 2012.
[6] E. Ozyilmaz, N. Kokturk, G. Teksut, and T. Tatlicioglu, “Unsus-pected risk factors of frequent exacerbations requiring hospital admission in chronic obstructive pulmonary disease,”
Interna-tional Journal of Clinical Practice, vol. 67, no. 7, pp. 691–697, 2013.
[7] A. Banerjee, C. Koziol-White, and R. Panettieri Jr., “P38 MAPK inhibitors, IKK2 inhibitors, and TNF𝛼 inhibitors in COPD,”
Current Opinion in Pharmacology, vol. 12, no. 3, pp. 287–292,
2012.
[8] I. M. Adcock, K. F. Chung, G. Caramori, and K. Ito, “Kinase inhibitors and airway inflammation,” European Journal of
Phar-macology, vol. 533, no. 1–3, pp. 118–132, 2006.
[9] P. J. Barnes, “Novel signal transduction modulators for the treatment of airway diseases,” Pharmacology and Therapeutics, vol. 109, no. 1-2, pp. 238–245, 2006.
[10] S. Rajendrasozhan, S. Chung, I. K. Sundar, H. Yao, and I. Rahman, “Targeted disruption of NF-𝜅B1 (p50) augments cigarette smoke-induced lung inflammation and emphysema in mice: a critical role of p50 in chromatin remodeling,” The
American Journal of Physiology—Lung Cellular and Molecular Physiology, vol. 298, no. 2, pp. L197–L209, 2010.
[11] U. Pannicke, B. Baumann, S. Fuchs et al., “Deficiency of innate and acquired immunity caused by an IKBKB mutation,” The
New England Journal of Medicine, vol. 369, no. 26, pp. 2504–
2514, 2013.
[12] A. Page, M. Navarro, M. Gar´ın et al., “IKKΒ leads to an inflammatory skin disease resembling interface dermatitis,”
Journal of Investigative Dermatology, vol. 130, no. 6, pp. 1598–
1610, 2010.
[13] G. Hu, A. Y. Gong, A. L. Roth et al., “Release of luminal exo-somes contributes to TLR4-mediated epithelial antimicrobial defense,” PLoS Pathogens, vol. 9, no. 4, Article ID e1003261, 2013. [14] C. Zaph, A. E. Troy, B. C. Taylor et al., “Epithelial-cell-intrinsic IKK-𝛽 expression regulates intestinal immune homeostasis,”
Nature, vol. 446, no. 7135, pp. 552–556, 2007.
[15] A. Tsuchiya, K. Imai, K. Asamitsu, Y. Waguri-Nagaya, T. Otsuka, and T. Okamoto, “Inhibition of inflammatory cytokine production from rheumatoid synovial fibroblasts by a novel I𝜅B kinase inhibitor,” Journal of Pharmacology and Experimental
Therapeutics, vol. 333, no. 1, pp. 236–243, 2010.
[16] K. R. Aupperle, B. L. Bennett, Z. Han, D. L. Boyle, A. M. Manning, and G. S. Firestein, “NF-𝜅B regulation by I𝜅B kinase-2 in rheumatoid arthritis synoviocytes,” Journal of Immunology, vol. 166, no. 4, pp. 2705–2711, 2001.
[17] H. H. Salem, B. Trojanowski, K. Fiedler et al., “Long-term IKK2/NF-𝜅B signaling in pancreatic 𝛽-cells induces immune-mediated diabetes mellitus,” Diabetes, vol. 63, no. 3, pp. 960– 975, 2014.
[18] S. Rajendrasozhan, J. Hwang, H. Yao, N. Kishore, and I. Rahman, “Anti-inflammatory effect of a selective I𝜅B kinase-beta inhibitor in rat lung in response to LPS and cigarette smoke,” Pulmonary Pharmacology and Therapeutics, vol. 23, no. 3, pp. 172–181, 2010.
[19] V. V. Kravchenko, J. C. Mathison, K. Schwamborn, F. Mercurio, and R. J. Ulevitch, “IKKi/IKK𝜀 plays a key role in integrating signals induced by pro-inflammatory stimuli,” The Journal of
Biological Chemistry, vol. 278, no. 29, pp. 26612–26619, 2003.
[20] C. Y. Chen, “A novel integrated framework and improved methodology of computer-aided drug design,” Current Topics
in Medicinal Chemistry, vol. 13, no. 9, pp. 965–988, 2013.
[21] K. C. Chen, S. S. Chang, F. J. Tsai, and C. Y. Chen, “Han ethnicity-specific type 2 diabetic treatment from traditional Chinese medicine?” Journal of Biomolecular Structure and
Dynamics, vol. 31, no. 11, pp. 1219–1235, 2013.
[22] Y.-T. Chang, W.-D. Lin, Z.-N. Chin et al., “Nonketotic hyper-glycinemia: a case report and brief review,” BioMedicine, vol. 2, no. 2, pp. 80–82, 2012.
[23] W.-L. Liao and F.-J. Tsai, “Personalized medicine: a paradigm shift in healthcare,” BioMedicine, vol. 3, no. 2, pp. 66–72, 2013. [24] D.-Y. Lin, F.-J. Tsai, C.-H. Tsai, and C.-Y. Huang, “Mechanisms
governing the protective effect of 17𝛽-estradiol and estrogen receptors against cardiomyocyte injury,” BioMedicine, vol. 1, no. 1, pp. 21–28, 2011.
[25] W.-Y. Lin, H.-P. Liu, J.-S. Chang et al., “Genetic variations within the PSORS1 region affect Kawasaki disease development and coronary artery aneurysm formation,” BioMedicine, vol. 3, no. 2, pp. 73–81, 2013.
[26] F.-J. Tsai, “Biomedicine brings the future nearer,” BioMedicine, vol. 1, no. 1, p. 1, 2011.
[27] F.-J. Tsai, “Rare diseases: a mysterious puzzle,” BioMedicine, vol. 3, no. 2, p. 65, 2013.
[28] I. C. Chou, W.-D. Lin, C.-H. Wang et al., “M¨obius syndrome in a male with XX/XY mosaicism,” BioMedicine, vol. 3, no. 2, pp. 102–104, 2013.
[29] I. C. Chou, W.-D. Lin, C.-H. Wang et al., “Association analysis between Tourette's syndrome and two dopamine genes (DAT1, DBH) in Taiwanese children,” BioMedicine, vol. 3, no. 2, pp. 88– 91, 2013.
[30] C.-C. Lee, C.-H. Tsai, L. Wan et al., “Increased incidence of
Parkinsonism among Chinese with𝛽-glucosidase mutation in
central Taiwan,” BioMedicine, vol. 3, no. 2, pp. 92–94, 2013. [31] C.-H. Wang, W.-D. Lin, D.-T. Bau, I. Chou, C. Tsai, and F.
Tsai, “Appearance of acanthosis nigricans may precede obesity: an involvement of the insulin/IGF receptor signaling pathway,”
BioMedicine, vol. 3, no. 2, pp. 82–87, 2013.
[32] C.-H. Wang, W.-D. Lin, and F.-J. Tsai, “Craniofacial dysmor-phism, what is your diagnosis?” BioMedicine, vol. 2, no. 2, pp. 49–50, 2012.
[33] T. Y. Tsai, K. W. Chang, and C. Y. C. Chen, “IScreen: world's first cloud-computing web server for virtual screening and de novo drug design based on TCM database@Taiwan,” Journal of
Computer-Aided Molecular Design, vol. 25, no. 6, pp. 525–531,
2011.
[34] S. C. Yang, S. S. Chang, H. Y. Chen, and C. Y. Chen, “Identifica-tion of potent EGFR inhibitors from TCM Database@Taiwan,”
PLoS Computational Biology, vol. 7, no. 10, Article ID e1002189,
2011.
[35] T. T. Chang, M. F. Sun, H. Y. Chen et al., “Screening from the world's largest TCM database against H1N1 virus,” Journal of
Biomolecular Structure and Dynamics, vol. 28, no. 5, pp. 773–
786, 2011.
[36] K.-C. Chen and C. Y. C. Chen, “Stroke prevention by tradi-tional Chinese medicine? A genetic algorithm, support vector machine and molecular dynamics approach,” Soft Matter, vol. 7, no. 8, pp. 4001–4008, 2011.
[37] S. S. Chang, H. J. Huang, and C. Y. Chen, “Two birds with one stone? Possible dual-targeting H1N1 inhibitors from traditional Chinese medicine,” PLoS Computational Biology, vol. 7, no. 12, Article ID e1002315, 2011.
[38] K. C. Chen, M. F. Sun, S. C. Yang et al., “Investigation into potent inflammation inhibitors from traditional Chinese medicine,”
Chemical Biology and Drug Design, vol. 78, no. 4, pp. 679–688,
2011.
[39] C. Y. Chen, “TCM Database@Taiwan: the world's largest tradi-tional Chinese medicine database for drug screening in silico,”
PLoS ONE, vol. 6, no. 1, Article ID e15939, 2011.
[40] S. Yang, S. Chang, and C. Y. Chen, “Identifying HER2 inhibitors from natural products database,” PLoS ONE, vol. 6, no. 12, Article ID e28793, 2011.
[41] H. J. Huang, K. J. Lee, H. W. Yu et al., “Structure-based and ligand-based drug design for HER 2 receptor,” Journal of
Biomolecular Structure and Dynamics, vol. 28, no. 1, pp. 23–37,
2010.
[42] W. I. Tou, S. S. Chang, C. C. Lee, and C. Y. Chen, “Drug design for neuropathic pain regulation from traditional Chinese medicine,” Scientific Reports, vol. 3, p. 844, 2013.
[43] C. Y. Chen, Y. H. Chang, D. T. Bau et al., “Ligand-based dual target drug design for H1N1: swine flu—a preliminary first study,” Journal of Biomolecular Structure and Dynamics, vol. 27, no. 2, pp. 171–178, 2009.
[44] H. J. Huang and C. Y. Chen, “A possible solution for hair loss by inhibiting corticotropin-releasing factor (CRF) receptor from traditional Chinese medicine,” Journal of Biomolecular Structure
and Dynamics, 2013.
[45] H. Y. Chen, S. S. Chang, Y. C. Chan, and C. Y. Chen, “Discovery of novel insomnia leads from screening traditional Chinese medicine database,” Journal of Biomolecular Structure and
Dynamics, vol. 32, no. 5, pp. 776–791, 2014.
[46] H. J. Huang, Y. R. Jian, and C. Y. Chen, “Traditional Chinese medicine application in HIV: an in silico study,” Journal of
Biomolecular Structure and Dynamics, vol. 32, no. 1, pp. 1–12,
2014.
[47] I. C. Hung, S. S. Chang, P. C. Chang, C. C. Lee, and C. Y. Chen, “Memory enhancement by traditional Chinese medicine?”
Journal of Biomolecular Structure and Dynamics, vol. 31, no. 12,
pp. 1411–1439, 2013.
[48] K. C. Chen, Y. R. Jian, M. F. Sun, T. T. Chang, C. C. Lee, and C. Y. Chen, “Investigation of silent information regulator 1 (Sirt1) agonists from traditional Chinese medicine,” Journal of
Biomolecular Structure and Dynamics, vol. 31, no. 11, pp. 1207–
1218, 2013.
[49] Y. A. Tsou, K. C. Chen, H. C. Lin, S. S. Chang, and C. Y. Chen, “Uroporphyrinogen decarboxylase as a potential target for specific components of traditional Chinese medicine: a virtual screening and molecular dynamics study,” PLoS ONE, vol. 7, no. 11, Article ID e50087, 2012.
[50] K. Y. Chen, S. S. Chang, and C. Y. Chen, “In silico identification of potent pancreatic triacylglycerol lipase inhibitors from tra-ditional Chinese medicine,” PLoS ONE, vol. 7, no. 9, Article ID e43932, 2012.
[51] H.-J. Huang, C.-C. Lee, and C. Y.-C. Chen, “Pharmacological chaperone design for reducing risk factor of Parkinson’s disease from traditional Chinese medicine,” Evidence-Based
Comple-mentary and Alternative Medicine, vol. 2014, Article ID 830490,
12 pages, 2014.
[52] K. W. Chang, T. Y. Tsai, K. C. Chen et al., “iSMART: an integrated cloud computing web server for traditional Chinese
medicine for online virtual screening, de novo evolution and drug design,” Journal of Biomolecular Structure and Dynamics, vol. 29, no. 1, pp. 243–250, 2011.
[53] S. Liu, Y. R. Misquitta, A. Olland et al., “Crystal structure of a human I𝜅B Kinase 𝛽 asymmetric dimer,” The Journal of
Biological Chemistry, vol. 288, no. 31, pp. 22758–22767, 2013.
[54] C. Y. C. Chen and W. L. Tou, “How to design a drug for the disordered proteins?” Drug Discovery Today, vol. 18, no. 19-20, pp. 910–915, 2013.
[55] V. Zoete, M. A. Cuendet, A. Grosdidier, and O. Michielin, “SwissParam: a fast force field generation tool for small organic molecules,” Journal of Computational Chemistry, vol. 32, no. 11, pp. 2359–2368, 2011.
[56] U. D. Priyakumar and A. D. MacKerell, “Comparison of the CHARMM27, AMBER4. 1 and BMS nucleic acid force fields via free energy calculations of base flipping,” Abstracts of Papers of
the American Chemical Society, vol. 230, pp. U1391–U1392, 2005.
[57] B. Hess, C. Kutzner, D. van der Spoel, and E. Lindahl, “GRGMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation,” Journal of Chemical Theory
and Computation, vol. 4, no. 3, pp. 435–447, 2008.
[58] W. L. Tou and C. Y. Chen, “May disordered protein cause serious drug side effect?” Drug Discovery Today, vol. 19, no. 4, pp. 367– 372, 2014.
[59] T. A. Darden and L. G. Pedersen, “Molecular modeling: an experimental tool,” Environmental Health Perspectives, vol. 101, no. 5, pp. 410–412, 1993.
[60] M. Yoshikawa, S. Hatakeyama, Y. Inoue, and J. Yamahara, “Saussureamines A, B, C, D, and E, new anti-ulcer principles from Chinese Saussureae radix,” Chemical and Pharmaceutical
Bulletin, vol. 41, no. 1, pp. 214–216, 1993.
[61] T. Nakazawa, T. Yasuda, and K. Ohsawa, “Metabolites of orally administered Magnolia officinalis extract in rats and man and its antidepressant-like effects in mice,” Journal of Pharmacy and
Pharmacology, vol. 55, no. 11, pp. 1583–1591, 2003.
[62] Y. K. Choi, S. G. Cho, S. M. Woo et al., “Saussurea lappa Clarke-derived costunolide prevents TNF𝛼-induced breast cancer cell migration and invasion by inhibiting NF-𝜅B activity,”
Evidence-Based Complementary and Alternative Medicine, vol. 2013,
Article ID 936257, 10 pages, 2013.
[63] J. M. Walker, A. Maitra, J. Walker, M. M. Ehrnhoefer-Ressler, T. Inui, and V. Somoza, “Identification of Magnolia officinalis L. Bark extract as the most potent anti-inflammatory of four plant extracts,” The American Journal of Chinese Medicine, vol. 41, no. 3, pp. 531–544, 2013.
[64] J. Y. Xie, H. Y. Di, H. Li, X. Cheng, Y. Zhang, and D. Chen, “Bupleurum chinense DC polysaccharides attenuates lipopolysaccharide-induced acute lung injury in mice,”
Phy-tomedicine, vol. 19, no. 2, pp. 130–137, 2012.
[65] S. N. Park, Y. K. Lim, M. O. Freire, E. Cho, D. Jin, and J.
Kook, “Antimicrobial effect of linalool and𝛼-terpineol against
periodontopathic and cariogenic bacteria,” Anaerobe, vol. 18, no. 3, pp. 369–372, 2012.
[66] R. A. Laskowski and M. B. Swindells, “LigPlot+: multiple ligand-protein interaction diagrams for drug discovery,” Journal of
Chemical Information and Modeling, vol. 51, no. 10, pp. 2778–
Submit your manuscripts at
http://www.hindawi.com
Stem Cells
International
Hindawi Publishing Corporation
http://www.hindawi.com Volume 2014
Hindawi Publishing Corporation
http://www.hindawi.com Volume 2014 INFLAMMATION
Hindawi Publishing Corporation
http://www.hindawi.com Volume 2014
Behavioural
Neurology
Endocrinology
International Journal ofHindawi Publishing Corporation
http://www.hindawi.com Volume 2014
Hindawi Publishing Corporation
http://www.hindawi.com Volume 2014
Disease Markers
Hindawi Publishing Corporation
http://www.hindawi.com Volume 2014
BioMed
Research International
Oncology
Journal ofHindawi Publishing Corporation
http://www.hindawi.com Volume 2014
Hindawi Publishing Corporation
http://www.hindawi.com Volume 2014
Oxidative Medicine and Cellular Longevity
Hindawi Publishing Corporation
http://www.hindawi.com Volume 2014
PPAR Research
The Scientific
World Journal
Hindawi Publishing Corporation
http://www.hindawi.com Volume 2014
Immunology Research
Hindawi Publishing Corporation
http://www.hindawi.com Volume 2014
Journal of
Obesity
Journal ofHindawi Publishing Corporation
http://www.hindawi.com Volume 2014
Hindawi Publishing Corporation
http://www.hindawi.com Volume 2014
Computational and Mathematical Methods in Medicine
Ophthalmology
Journal of Hindawi Publishing Corporationhttp://www.hindawi.com Volume 2014
Diabetes Research
Journal of Hindawi Publishing Corporationhttp://www.hindawi.com Volume 2014
Hindawi Publishing Corporation
http://www.hindawi.com Volume 2014
Research and Treatment
AIDS
Hindawi Publishing Corporation
http://www.hindawi.com Volume 2014
Gastroenterology Research and Practice
Hindawi Publishing Corporation
http://www.hindawi.com Volume 2014