National Chiao-Tung University PhD Thesis
國立交通大學博士學位論文
Functional study of the phosphodiesterase YjcC and the second
messenger cyclic di-GMP in the stress response regulation in
Klebsiella pneumoniae CG43
克雷白氏肺炎桿菌 CG43 磷酸二酯酶 YjcC 與二級訊息分子 c-di-GMP
在壓力反應調控之功能性研究
分子醫學及生物工程所
學生:黃靜柔
Student: Ching-Jou Huang
指導教授:彭慧玲 博士
Advisor: Hwei-Ling Peng,Ph.D.
中華民國一
○二年七月
II
克雷白氏肺炎桿菌 CG43 磷酸二酯酶 YjcC 與二級訊息分子 c-di-GMP
在壓力反應調控之功能性研究
Functional study of the phosphodiesterase YjcC and the second
messenger cyclic di-GMP in the stress response regulation in
Klebsiella pneumoniae CG43
研究生: 黃靜柔 Student: Ching-Jou Huang
指導教授: 彭慧玲 Advisor: Hwei-Ling Peng
國立交通大學
分子醫學及生物工程所
博士論文
National Chiao Tung University
Institute of Molecular medicine and Bioengineering
PHD Thesis
July 2013
III 謝誌 (Acknowledgement) 時間總是過得很快,轉眼來到交大已經七年了,博士班期間最感謝的就是我的 指導教授彭慧玲老師,很感謝老師不嫌棄我學術背景的薄弱,總是很有耐心的訓 練我獨立思考的能力,也常常在我學習過程中訓練並導正我的邏輯。雖然我自己 覺得走的很慢但還好老師願意陪我慢慢走,這是我始終非常感謝的。除了給予我 們的專業知識的訓練,老師也很關心我們的生活,這是一般教授不會做的事,所 以我真的由衷感謝我的彭老師,再多的感謝我也無法用言語或文字表達。 謝謝清華大學張晃猷老師,對於我的研究上,提醒我除了實驗結果的探討,必 需對自己所研究的相關新領域,都要有追根究底的態度,而不是只是走馬看花的 帶過去。很謝謝老師在我最後口試的時候給的一些建議,提醒自己,整理實驗結 果時除了主觀的相信自己所做的之外,還是要謹慎探討可能會出錯的部分。 感謝中興大學鄧文玲老師,在專業領域上給予我一些建議及幫助,也感謝老師 抽空參與了我在學期間重要的兩次口試,鄧老師熱心與直爽的個性,是我應該學 習的榜樣。雖然最後一次的論文口試,鄧老師沒有機會參與,但我還是很謝謝老 師寫信給我的鼓勵。 感謝林志生老師撥冗參與我在學期間的三次重要口試,雖然都是藉著口試的機 會接觸林老師,但可以很確定老師真的很關心我們將來出社會後,面對事情時, 應持有的正確態度。也很謝謝老師提醒我,外面的世界不像學校那麼單純,勉勵 我要繼續努力,努力不懈後才能嚐到甜的果實。 謝謝楊昀良老師總是很親切的給予我指導與建議,老師和善的態度也讓非常容 易緊張且自認有上台恐慌症的我,口試時的壓力減輕不少,雖然我的畢業口試沒 能邀請到楊老師,但是也是很謝謝老師參與我的資格考以及非論文口試。 謝謝中國醫藥大學林靖婷老師,林老師正好參與我的非論文及畢業口試論文, 雖然曾是實驗室的學姐,但隨著時間的累積,也讓我見識到靖婷學姐努力耕耘後 的成績。這是我們做為學弟妹所該學習的榜樣。也謝謝梁美智老師參與我最後畢 業論文的口試,梁老師提醒了我在表現口試的答問上,須考慮如何才能以條理分 明的方式,把自己的研究充分的表現出來。感謝過去彭老師實驗室眾多的學長姐
IV 及學弟妹。首先是丸子,盈蓉學姐,當我實驗遇到瓶頸或生活上有煩惱時,總是 帶著我一起去跳街舞,排解當時的煩惱與壓力,是我的精神支柱之一。謝謝健誠 與小新,在我博士班修讀期間,兩位彭家大學長總是能一針見血且精闢的指正我 們的實驗設計可能會遇到的缺失,也給予我很多的建議。除此之外,也很感謝健 誠在離開交大後,當我遇到問題,想尋求同儕意見時,樂意提供了一些建議或幫 忙,也常給我精神打氣。謝謝哲充學弟,在我博班後期,我們還一起合作把文章 搞定。 感謝在我博士班前幾年一起打拼的學弟妹,特別是雅雯、家華、秉熹、顗峰、 佩君、蕙如,謝謝你們總是很關心我的健康,也常拿好吃的零食、補品還有出國 買回來送給我的禮物,都是支持我一步一步往前走的動力,真的很謝謝你們。還 要感謝可愛學弟妹,品瑄、崴云、小波、力成、郁勝,總是默默的鼓勵我繼續往 前。最後要感謝陪伴我在博班後期時的學弟妹與生力軍:補教界未來名師:冠男、 多才多藝的正妹燕曦、講義氣的子祥、細心個性好的偉豐、可愛且聲音爽朗的珍 儀、溫柔體貼的俐君、閩南語天后兼彭家主計處處長蕙瑜、思慮清楚的專題生豐 碩,非常感謝你們在我最後要畢業時,所帶給我的歡笑以及鼓勵。新彭家生力軍 碩班學弟妹瑋芝、家睿、專題生煥義,期待大家能在彭家一起茁壯。 最後我要感謝我的爸媽,一直給我很大的鼓勵及讓我無後顧之憂,父母對我的 愛是無人能及的。也很感謝我的大姐、大姐夫、二姐、二姐夫以及小妹,一直以 來都代替我陪伴媽媽與爸爸,家人給予我的支持才讓我勇敢的走下去。還要感謝 台北三重的家人,包括大阿姨、小阿姨、小姨丈、兩個表妹依倫與李盈,更要感 謝從小就把我跟二姐帶大,已經在天上的三重阿嬤,很遺憾,在妳生前沒來的及 把喜悅帶給妳,我想阿嬤現在一定知道,也很開心我畢業了。謝謝北投三姨、三 姨丈給我的鼓勵及關心。感謝所有參與我生命過程的妳(你)們,才成就了現在的 我。
V 論文摘要
克雷白氏肺炎桿菌是一株伺機性的革蘭氏陰性病原菌,可造成肺炎、原發性
肝膿瘍、尿道和化膿性感染、及敗血症等廣泛疾病。我們實驗室先前利用小鼠活
體表現技術(in vivo expression technology),由克雷白氏肺炎桿菌 CG43 基因庫
中分離出只在小鼠體內表現、影響其毒性且會受巴拉刮(1,1'-二甲基-4,4'-聯吡 啶氯化物)誘導表現的 yjcC 基因,分析此基因序列顯示此具 534 個殘基的蛋白 質 N 端具有 2 個穿膜的二級結構和可能接收訊息的 CSS 功能區,其 C 端具有磷 酸二酯酶(phosphodiesterase; PDE)活性的功能區。本論文的第一部份,我們證 實 YjcC 可受巴拉刮和過氧化氫誘導表現;剔除 yjcC 基因除了使 CG43 抗氧化壓 力的能力減低外,也降低其莢膜多醣的合成及對小鼠的毒性;而 yjcC 基因缺損 株中的活性氧自由基、氧化傷害化、第三型線毛單位蛋白 MrkA 表現及生物膜活 性相對增加;同時,經純化的 YjcC 磷酸二酯酶功能區重組蛋白被證實具有 PDE
活性;進一步,在 CG43 增加 YjcC 表現使 c-di-GMP 濃度降低後的 mRNA 定序
結果顯示有 34 與 29 個基因分別因而提高和降低其表現量,而其中包括與氧化壓
力相關的基因。這些結果顯示 YjcC 在氧化壓力反應中扮演正向調控的角色(詳
述於第二章)。
細 菌 的 二 級 訊 息 分 子 cyclic di-GMP ( c-di-GMP ) 由 雙 鳥 苷 酸 環 化 酶
(diguanylate cyclase; DGC)環化合成,被磷酸二酯酶所分解,因此,c-di-GMP
VI
傳遞系統主導許多基因的表現與生理功能的調控。為了進一步探討克雷白氏肺炎
桿菌中 c-di-GMP 濃度是否參與調控細菌對抗各種壓力的反應,我們將具有 DGC
活性的質體 pRK415-ydeH 轉殖至 K. pneumoniae CG43S3 提高 c-di-GMP 的濃度
後,再以轉錄體 RNA 定序與定量分析受影響的基因,結果顯示除了已被證實受
c-di-GMP 濃度正向調控的基因 mrkABCDF 和 mrkHI 表現量明顯升高外,熱休克
反應及其它壓力反應相關的基因 ibpA、clpB、dnaK、grxA 和 dinI 也大量表現; 而比較 K. pneumoniae CG43S3[pRK415]和 CG43S3[pRK415-ydeH]發現細胞內的 c-di-GMP 濃度上升使細菌抵抗 50℃的熱休克反應的能力增加,相對的卻降低細 菌對抗氧化及酸壓力,此結果暗示細菌可藉改變 c-di-GMP 濃度來調控抵抗惡劣 環境的反應(詳述於第三章)。 K. pneumoniae CG43 基因體已於今年初定序完成,序列分析結果顯示至少有 25 個與 c-di-GMP 濃度調節相關的基因,釐清這些基因的表現調控和功能性,將 有助於建立 c-di-GMP 主導調控的基因網路,也將有利於了解 c-di-GMP 在細菌 對抗壓力反應的調控機制。另外,最近研究證實 c-di-GMP 可與 STING 蛋白結合 而增強人類干擾素的表現,又因其增強免疫反應的特性,可提供作為疫苗佐劑; 未來,如何應用 c-di-GMP 配合抗生素的使用來降低細菌的感染將是一個值得深 入探討的課題。
VII
Thesis abstract
Klebsiella pneumoniae is an opportunistic Gram-negative pathogen that causes a
wide range of infections, including pneumonia, urinary tract, purulent infections,
primary liver abscess and septicemia. We have previously identified from K.
pneumoniae CG43, using in vivo expression technology (IVET), yjcC gene which was
shown to be inducible by paraquat and affect its virulence to mouse. Sequence
analysis of YjcC shows a signal peptide followed by 2 transmembrane domains and a
CSS motif at the N-terminal region, whereas the C-terminal contains a conserved
EAL domain of the PDE enzyme. For the first part of the thesis, we have
demonstrated that yjcC is induced expression by paraquat and H2O2. The yjcC
deletion reduced the bacterial oxidative stress resistant activity, capsular
polysaccharide production, and virulence to mouse. In addition, the yjcC deletion
mutant exhibited increased production of reactive oxygen species, oxidative damage,
type 3 fimbriae MrkA pilin, and biofilm. The recombinant protein containing the
YjcC-EAL domain was demonstrated to exhibit phosphodiesterase (PDE) activity.
Moreover, transcriptome analysis via RNAseq of CG43S3[pRK415-yjcC] compared
to the CG43[pRK415] gene expression revealed 34 upregulated and 29 downregulated
genes, which include stress related genes. The results suggest that yjcC plays a
VIII (Detailed description in Chapter 2).
In bacteria, the second messenger c-di-GMP is regulated by diguanylate cyclase
(DGC) enzymes and phosphodiesterases (PDEs) that catalyze synthesis and
hydrolysis of this molecule, respectively. Many recent reports show that the
c-di-GMP-mediated signal transduction system is a major regulator for many gene
expression and physiological response. In order to investigate if c-di-GMP level is
involved in the stress response regulation in K. pneumoniae, the DGC expression
plasmid pRK415-ydeH was used to transform K. pneumoniae CG43S3 to elevate the
intracellular c-di-GMP level. Subsequently, transcriptome analysis via RNAseq was
employed and then qRT-PCR analysis used to confirm the affected genes. The results
showed that, in addition to the reported c-di-GMP upregulated genes mrkABCDF and
mrkHI, the heat shock response and other stress response genes including ibpA, clpB,
dnaK, grxA, and dinI are also increasingly expressed. Compared to CG43S3[pRK415],
CG43S3[pRK415-ydeH] had increased the heat shock (50℃) resistant activity, but
had reduced the bacterial resistance to oxidative and acid stress by the increase of the
c-di-GMP levels. The results imply that bacteria could resist to the harsh environment
by modulating the intracellular c-di-GMP levels.
Analysis of the recently resolved genome sequence of K. pneumonia CG43
IX
for the intracellular c-di-GMP level modulation. To clarify how these gene expression
are regulated and what are their functional roles in the bacteria should help to
establish a c-di-GMP-dependent gene network. These data should also provide much
more insights of the c-di-GMP role in the stress response regulation. Besides, several
recent reports have demonstrated that c-di-GMP is able to bind STING protein, the
stimulator of interferon gene, thereby enhances the immune response. It is proposed to
be used as vaccine adjuvant because of the potent immune modulator property. In the
future, the issue of how to apply c-di-GMP together with antibiotic treatment to
X
Table of Contents
謝誌 ... Ⅲ 論文摘要(Thesis Abstract in Chinese) ... Ⅴ Thesis Abstract... Ⅶ Table of Contents ... Ⅹ List of Tables ... ⅩⅡ List of Figures ... ⅩⅢ Abbreviations ... ……..ⅩⅣ Chapter 1 General Introduction ... 1 1.1 Klebsiella pneumoniae ... 2 1.1.1 K. pneumoniae infections... 2
1.2 K. pneumoniae virulence factors... 4
1.2.1 K. pneumoniae type 1 fimbriae ... 6
1.2.2 K. pneumoniae type 3 fimbriae ... 6
1.3 Cyclic-di-GMP signaling system ... 7
1.3.1 The GGDEF- and EAL domain proteins ... 8
1.3.2 Regulatory mechanism of c-di-GMP ... 9
1.3.3 Role of c-di-GMP in virulence ... 10
1.3.4 Domain structure of GGDEF- and EAL- domains in K. pneumoniae CG43S3 ... 11
1.4 Oxidative stress ... 13
1.4.1 Oxidative stress response in bacteria ... 14
XI
1.5.1 RPKM measure ... 18
Chapter 2 YjcC, a c-di-GMP phosphodiesterase protein, regulates the oxidative stress response and virulence of Klebsiella pneumoniae CG43 2.1. Abstract ... 20
2.2. Introduction ... 21
2.3. Results ... 23
2.4. Discussion ... 29
Chapter 3 Transcriptome analyses of the c-di-GMP effect on the stress response in Klebsiella pneumoniae CG43S3 3.1 Abstract ... 49 3.2 Introduction ... 49 3.3 Results ... 50 3.4 Discussion ... 52 Chapter 4 Conclusion and Perspectives... 60
Chapter 5 Experimental Section ... 65
References ... 82
Publication ... 102
XII
List of Tables
Table 2.1. YjcC effect on the mouse virulence ... 34
Table 2.2. Significantly upregulated genes by yjcC overexpression ... 35
Table 2.3. Significantly downregulated genes by yjcC overexpression ... 36
Table 3.1. Upregulated genes from the comparison of CG43S3[pRK415-ydeH]
and CG43S3[pRK415] ... 53
Table 3.2. Downregulated genes from the comparison of CG43S3[pRK415-ydeH]
and CG43S3[pRK415] ... 54
Table 5.1. Strains and plasmids used in this study ... 80
XIII
List of Figures
Fig. 1.1 Domain architecture of putative c-di-GMP signaling proteins encoded of K.
pneumoniae CG43 ... 12
Fig. 2.1 yjcC gene is paraquat inducible, and SoxRS and RpoS dependent ... 38
Fig. 2.2 Analysis of the deletion effects of yjcC upon exposure to oxidative stress ... 40
Fig. 2.3 Deletion of yjcC places bacteria in an oxidative stress state ... 42
Fig. 2.4 YjcC affects the CPS biosynthesis, biofilm formation and MrkA production ... 45
Fig. 2.5 The qRT-PCR analysis of the expression of mrkA, mrkH, and mrkI of K. pneumoniae CG43S3 ... 47
Fig. 3.1 The cyclic di-GMP content... 56
Fig. 3.2 qRT-PCR analysis of the selected up-regulated genes ... 57
Fig. 3.3 Heat shock stress survival analysis ... 58
XIV
Abbreviations
PLA pyogenic liver abscess
ESBL extended-spectrum β-lactamase
CPS capsular polysaccharide
LPS lipopolysaccharide
C-di-GMP Bis-(3’-5’)-cyclic dimeric guanosine monophosphate
DGC di-guanylate cyclase
PDE phosphodiesterase
qRT-PCR quantitative real-time polymerase chain reaction EDTA ethylenediaminetetraacetic acid
CFU colony forming unit
IPTG isopropyl 1-thio-β-D-galactopyranoside
OD optical density
1
Chapter 1
2
1.1 Klebsiella pneumoniae
Klebsiella pneumoniae is a rod-shaped, non-motile, heavily-encapsulated
gram-negative bacterium belonging to the Enterobacteriaceae. As an opportunistic
human nosocomial pathogen, it is primarily attacks immunocompromised individuals.
Mostly community-acquired K pneumoniae infections cause pneumonia or urinary
tract infections [1]. Since 1980s, liver abscesses patients infected with K. pneumoniae
were start to be described in reports and case series in Taiwan [2,3].
1.1.1K. pneumoniae infections
K. pneumoniae has emerged as the leading cause of pyogenic liver abscess in Asia,
with over 900 cases reported in Taiwan by 2004 [4]. Smaller series have been
reported from other countries in Asia, including the Peoples Republic of China, Korea,
Japan, Singapore, Hong Kong, Thailand and India [5]. Diabetes mellitus or impaired
glucose tolerance is significant comorbidity, present in 40–75 % of these cases. A
significant minority of patients (8–15%) had infection at other anatomical sites,
including brain abscess, endophthalmitis, pyogenic meningitis, empyema, septic
pulmonary emboli, osteomyelitis, septic arthritis, prostatic and psoas abscesses.
Mortality ranged from 4 to 11 %, which is notably lower than historical mortality
rates reported for pyogenic liver abscesses [6,7]. In 1999, K. pneumoniae was first
3
in North America [6,8]. In Europe, single cases have been reported from Spain,
Belgium, the Netherlands, Sweden, Italy and France [9-13]. One larger case series
from France published in 2011 detailed seven cases of invasive liver abscess from
hospital [14]. In addition, it was identified as the commonest cause (23/79 cases) of
pyogenic liver abscess at two New York City hospitals, between 1993 and 2003 [15].
Invasive isolates typically belong to capsular serotypes K1 and K2, both of which
express a distinct hypermucoviscous phenotype [16]. Heavy encapsulation confers the
bacteria resistance to phagocytosis and prevent from intracellular killing. Two
plasmid-encoded virulence factors have been well characterized, rmpA, a regulator of
mucoid phenotype that upregulates the capsule synthesis, and the iron siderophore
aerobactin, which enables the bacterium to obtain iron. Other virulence factors
include the genes kfu, which codes for an iron uptake system, and allS, which is
associated with allantoin metabolism. All have been associated with severe pyogenic
infections [17,18].
In the hospital environment with the extensive use of antibiotics, multiple drug
resistance has been increasingly observed in K. pneumoniae, especially the
extended-spectrum β-lactamase (ESBL)-producing strains [19-21]. Carbapenems are
considered to be the preferred agents for the treatment of serious infections caused by
4
hydrolysis and observed retained susceptibility of ESBL producers [22-24]. Nerveless,
it is rare occurrence for liver abscesses caused by ESBL- K pneumoniae have been
reported in Taiwan. Carbapenem-resistant K pneumoniae, such as strains producing
NDM-1, has been increasingly reported [25,26]. Antibiotics such as
ampicillin–sulbactam, a third-generation cephalosporin, aztreonam, and a quinolone
can be used t treat these hyper-resistance strains [27].
1.2 Klebsiella pneumoniae virulence factors
The virulence factors that allow K. pneumoniae to overcome innate host immunity
and to maintain infection in a mammalian host factors include capsule,
lipopolysaccharides, adhesins, iron acquisition systems, serum resistance factor, and
biofilm formation regulators [28,29]. CPS acts to protect the bacteria from
phagocytosis, from killing by polymorphonuclear granulocytes and from killing by
bactericidal serum factors [30-32]. Besides the physical hindrance to fimbrial binding,
the role of Klebsiella CPS in mediating the bacterial resistance to antimicrobial
peptides has also been reported [33,34]. K. pneumoniae strains expressing K1 and K2
CPS are the most virulent to mice [35]. In addition to the 77 serotypes distinguished, a
new K serotype has been identified in 2008 [36]. Serotypes K1, K2, K4 and K5 are
highly virulent in experimental infection in mice and are often associated with severe
5
be the most prevalent capsular serotypes in liver abscess-causing K. pneumoniae. The
rmpA (regulator of the mucoid phenotype A) gene correlated with abscess formation
in patients with community-acquired K. pneumoniae bacteremia has been attributed to
be a risk factor for metastatic infection in patients with K. pneumoniae liver abscess
[38]. The rmpA together with rmpA2 gene both located on the large virulence plasmid
pLVPK are able to enhance the CPS biosynthesis thereby confer K. pneumoniae a
hypermucoviscosity phenotype [39].
Animal experiments have been performed to assess the role of iron-acquisition
systems in K. pneumoniae pathogenicity [40,41]. Analysis of the genomic sequence of
K. pneumoniae NTUH-K2044 revealed 10 putative iron-acquisition systems, whereas
K. pneumoniae strain MGH78578 and CG43 possess only 6 and 8 of these systems,
respectively [42].
The LPS O-antigen and the lipid A are responsible for the resistance to serum
factors and the establishment of septic shock [43]. Antimicrobial peptides, such as
polymyxin B, are bactericidal agents that exert their effects by interacting with the
LPS of Gram-negative bacteria. Our previous studied have shown that regulation of
the gene expression for LPS modification determine the polymyxin B resistance
[44,45]. The adhesion factors, including the type 1 [46,47] and 3 fimbriae [48] play
6 formation.
1.2.1 K. pneumoniae type 1 fimbriae
Type 1 fimbriae are approximately 7 nm wide and 1-2 μm long surface organelles
are found in many bacteria in the family Enterobacteriaceae [49,50]. Type 1 fimbriae
are encoded by fimAICDEFGH gene cluster coding for the fimbrial structure and
assembly. The fimbrial rod consists of the major subunits FimA and the minor
subunits FimI, FimF, and FimG. The adhesive properties of type 1 fimbriae are
exerted by the FimH adhesin which locates at the tips of the fimbriae. FimC and
FimD are respectively chaperone and usher that are required for the fimbrial assembly.
[51,52]. Type 1 fimbriae, which are able to cause mannose-sensitive agglutination of
yeast cells or erythrocytes (mannose-sensitive haemagglutination, MSHA) from
guinea pig are regulated via phase- variation [47,51,53-57].
The fimK gene, locating downstream to the fimH gene and unique to the K.
pneumoniae fim gene cluster, encodes an EAL domain protein [56]. We have recently
demonstrated that FimK may influence type 1 fimbriation by binding to fimS via the
N-terminal domain, and thereafter, the altered protein structure activates C-terminal
PDE activity to reduce the intracellular c-di-GMP level[58].
1.2.2 K. pneumoniae type 3 fimbriae
7
wide and 0.5-2 μm long surface organelles. The fimbriae are able to agglutinate the
tannic acid-treated human erythrocytes which could not be competitively inhibited by
mannose and hence designated MR/K haemagglutination [59]. Besides playing an
important role in biofilm formation on biotic and abiotic surfaces, type 3 fimbriae are
able to attach the endothelial and bladder epithelial cell lines [60-65]. The fimbriae
are encoded by chromosomally- or plasmid-borne mrkABCDF [61,65,66]. MrkD is
the adhesin that mediates binding specificity and biofilm formation on extracellular
matrix-coated surfaces , which can bind to type IV and/or type V collagen [63].
1.3 Cyclic-di-GMP signaling system
In bacteria, there are having various ways to sense environmental signals and to
adapt their behavior and physiology through different signaling transduction systems.
In bacteria, there are various ways to sense environmental signals and to adapt their
behavior and physiology through different signaling transduction systems. Cyclic
AMP (cAMP) is a global second messenger involved in bacterial transcription
regulation, while the signaling role of cyclic di-GMP (c-di-GMP) which was first
discovered in G. xylinus [67] as an allosteric activator of the cellulose synthesis has
not been recognized until recently. The second messenger c-di-GMP exclusively
found in bacteria is involved in many fundamental behaviors such as motility,
8
biofilm formation, and developmental transitions [68,69] [70].
1.3.1 The GGDEF- and EAL-domain proteins
In general, GGDEF domains are approximately 170 amino acids long and
GG(D/E)EF motif is an integral part of the active site [71]. Stand-alone GGDEF
domains are usually not enzymatically active, but require activation through an
N-terminal signaling domain for activation. Besides the EAL motif, there are several
other highly conserved motifs involved in catalysis, substrate binding and divalent ion
coordination. Usually, EAL domains show significant enzymatic activity without
N-terminal allosteric activation. Both GGDEF and EAL domain containing proteins
can be enzymatically active [72-74]. Alternatively, only one domain possesses
enzymatic activity, while the enzymatically inactive domain serves a regulatory
function [75].
GGDEF or EAL domain proteins often contain additional sensory and signal
transduction domains such as PAS, GAF, HAMP REC, and HTH domains [69,76,77].
It has been shown that oxygen, amino acids, electrons, and photons can modify the
activity of DGC or PDE proteins. For example, PAS is a conserved protein domain
involved in sensing oxygen, redox or light [78]; PleD is a GGDEF domain protein
that carries the REC domain that gets phosphorylated to activate the DGC activity for
9
Many GGDEF domain proteins possess c-di-GMP diguanyl cyclase (DGC)
activity, while EAL or HD-GYP domain proteins exert c-di-GMP phosphodiesterase
(PDE) activity [72,80,81]. The concentration of c-di-GMP is modulated through the
action of DGC and PDE respectively to synthesize and hydrolysis of c-di-GMP
[82,83].
1.3.2 Regulatory mechanism of c-di-GMP
Through binding to diverse receptors or effectors, the c-di-GMP exerts a regulatory
activity. The small ‘effector’ called PilZ domain, transcription factor or riboswitch
[84-86]. The c-di-GMP-mediated regulation can occur at the level of transcription,
post-transcription or post-translation, such as in the allosteric effect on cellulose
synthesis or regulation of protein turnover [70]. In general, c-di-GMP is involved in
positive regulation of exopolysaccharide production, biosynthesis of adhesive
fimbriae and biofilm formation [58,82,87-93]. By contrast, motility is commonly
negatively regulated by c-di-GMP. Various types of motility, including
flagella-mediated swimming and swarming, type IV pili-mediated twitching motility
are all affected by c-di-GMP [87,94]. In S. typhimurium, high levels of c-di-GMP
generated by overexpression of the DGC AdrA inhibited swarming and swimming
motility while reduction of c-di-GMP levels by overexpression of the PDE YhjH
10
YcgR leads to a conformational change in the protein and consequently the c-di-GMP
loaded YcgR form a complex with FliG and FliM to impair the flagella activity
[85,98,99].
1.3.3 Role of c-di-GMP in virulence
C-di-GMP signaling is involved in regulating virulence of many bacteria which
include E. coli, S. Typhimurium, Vibrio cholerae, Pseudomonas aeruginosa,
Bordetella pertussis, Xanthomonas campestris, Legionella pneumophila, Brucella
melitensis and Anaplasma phagocytophilum [75,93,98,100-105]. In V. cholera,
downregulation of c-di-GMP levels by the PDE VieA leads to activation of cholera
toxin [106]. P. aeruginosa c-di-GMP signaling is required for biofilm formation in
chronic infection and also the acute infection phenotype [90,107,108]. The plant
pathogen X. campestris pathovar campestris (Xcc) causes disease through a HD-GYP
domain protein expression to regulate the production of extracellular enzymes and
extracellular polysaccharide[109], and motility. In S. typhimurium, the EAL-domain
like protein STM1344 causes resistance to oxidative stress, inhibits rapid macrophage
killing and is required for virulence in the typhoid fever mouse model. Intriguingly,
STM1344 has no PDE activity nor has bind activity to c-di-GMP, and hence the
involvement of c-di-GMP signalling in these phenotypes remains to be investigated
11
1.3.4 Domain structure of GGDEF- and EAL-protein encoding genes in K.
pneumoniae CG43S3
GGDEF and EAL domain proteins are widespread in bacterial genomes. Very
often one bacterial genome contains more than one GGDEF and EAL domain protein.
The sequenced genome of S. typhimurium codes for 20 GGDEF/EAL domain proteins;
5 contain a GGDEF, 8 an EAL domain and 7 contain both. On the other hand, E. coli
K-12 has 12 GGDEF, 12 EAL and 7 GGDEF-EAL domain proteins [111]. A recent
study search for conserved GGDEF and EAL domains in three sequenced K.
pneumoniae genomes revealed multiple copies of GGDEF and EAL containing
proteins: 21 for K. pneumoniae Kp342, 18 for K. pneumoniae MGH 78578 and 17 for
K. pneumoniae NTUH-K2044 [112]. The K. pneumoniae CG43 genome sequence has
recently been resolved and the annotation results show 24 GGDEF- and EAL-domain
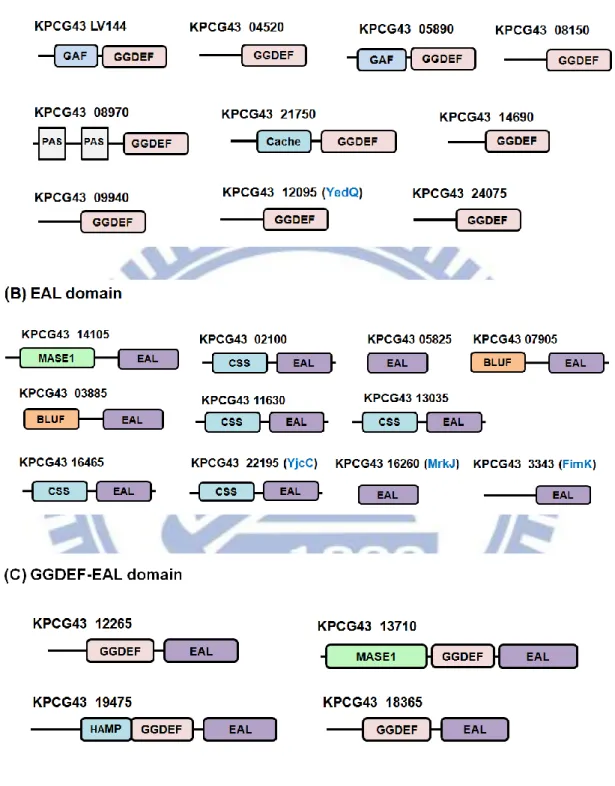
protein encoding genes. As shown in Fig. 1.1, 10 GGDEF-, 11 EAL- and 4
GGDEF-EAL-domain protein encoding genes are identified. The sensor domains
identified include MASE, CHASE, CACHE and CSS motif [113]. Genome-wide
approach to study these proteins might shed light on their functional roles in different
12
Fig. 1.1. Domain architecture of putative c-di-GMP signaling proteins encoded
by the K. pneumoniae CG43. The genome of K. pneumoniae CG43 encoding (A)
13
as indicated, was performed using the Pfam database provided online
(http://www.sanger.ac.uk /Software/Pfam/). The identities of the proteins are also
shown in blue: four EAL domain proteins (YjcC, FimK and MrkJ) that have been
described in K. pneumoniae [58,91,114,115] and (chapter 2).
1.4 Oxidative stress
The most commonly discussed oxidants that cause damage to DNA, proteins,
and cell membranes and often results in cell death are the reactive oxygen species
(ROS) including superoxide anion (O2.-), hydrogen peroxide (H2O2), and the hydroxyl
radical (HO.), and the reactive nitrogen species (RNS), which include nitric oxide
(NO.) and peroxynitrite (ONOO-). During infection, pathogens have equipped to
protect themselves from the oxidative burst of phagocytic cells and the challenging
oxidative environments within cellular and extracellular compartments.
The oxygen species can be excluded from active sites by electron transfer to the
redox cofactors. Reactions of this type occur by the formation of reactive oxygen
species (ROS) and their subsequent inactivation of enzymes. Intracellular molecular
oxygen can adventitiously abstract electrons from the exposed redox moieties of
electron-transfer enzymes, thereby generating partially reduced oxygen species. A
mixture of O2− and H2O2 is formed, reflecting the fact that either one or two electrons
14
abundant, in particular are responsible for all aerobic organisms to experience a
steady flux of endogenously generated oxidants [116]. The overall reaction rate is
proportional to collision frequency; thus, O2− and H2O2 fluxes depend directly upon
the ambient concentration of oxygen [117,118]. Several source of oxidative stress
have been identified that include (a) intracellular enzyme autooxidation. In
exponentially growing E.coli, both O2.-and H2O2 are generated by the auto-oxidation
of components of the respiratory chain [119]. (b) envirmental redox reactions, (c)
H2O2 released by competing microbes, (d) phagosomal NADPH oxidase and (e) redox
cycling antibiotics , plants or microorganisms secrete redox-cycling antibiotics that
diffuse into the competing bacteria, chemically oxidize redox enzymes and transfer
the electrons to molecular oxygen [118].
1.4.1 Oxidative stress response in bacteria
The defense mechanisms, which play an important role in determining the
bacterial virulence, include sensing, avoiding, and removing the oxidants. In
Escherichia coli, superoxide is removed by SODs (SodA, SodB, SodC) [120],
generating hydrogen peroxide which is then removed by catalases (KatE, KatG) and
peroxidases (AhpC). The transcriptome analysis of Pseudomonas aeruginosa or
15
virulence genes, the genes encoding products involved in DNA repair and anaerobic
metabolism were induced. Nevertheless, many of the genome-wide analysis revealed
the expression of more than 100 genes were induced in responding to either H2O2 or
paraquat indicating the complexity of the antioxidant strategies [121-123].
Many of the defenses are controlled by regulators that respond to iron (e.g., Fur),
oxygen tension (e.g., FNR and ArcAB), superoxide (e.g., SoxRS), and hydrogen
peroxide (e.g., OxyR). In E. coli, OxyR is known to regulate more than nine genes
including katG (hydroperoxidase I), ahpC (alkylhydroperoxide reductase), gorA
(gluthione reductase), oxyS (regulatory RNA), and fur that are involved directly or
indirectly in the oxidative stress response. The antioxidant genes katE, katG, and
sodC have been reported to be components of RpoS regulon .While the expression of
superoxide dismutase SodA and SodB are respectively controlled by at least 3 global
regulators including SoxRS, ArcAB, and Fur [124].
RpoS is a sigma factor which regulates expression of a variety of genes in both E.
coli and Salmonella spp. that allows the bacteria to adapt, resist, and survive under
stress condition. The deletion of rpoS in V. vulnificus was found to down-regulate the
expression of fur, indicating a cascade regulation between the two global regulators.
Fur, which is complexed with iron as an iron responsive regulator, binds to multiple
16
required for iron acquisition, acid and oxidative stress responses. However, Fur has
also been demonstrated to function as a transcriptional activator for several genes
including fur itself (Hp and Vv), and virulence associated genes [125-127].
In K. pneumoniae CG43, the 2CS response regulator encoding gene kvgA deletion
was found to reduce the expression of katG and sodC. KvgAS has also been shown to
be induced expression in upon treatment with 0.2 mM paraquat, which suggesting an
involvement of the 2CS in oxidative stress defense in the bacteria. Moreover, we have
reported in K. pneumoniae CG43 that rpoS deletion reduced the kvgAS expression
which also showing a cascade regulation on the expression of 2CS. This suggests
complex signaling networks with inter-connection regulatory circuits are required for
multiple stress signal integration [128].
1.5 Transcriptome analysis
Transcriptome analysis of gene expression can be profiled by high throughput
techniques including SAGE, serial analysis of gene expression [129], microarray
[130], and sequencing of clones from cDNA libraries [131]. Microarrays have been
the method of choice providing high throughput and affordable costs. However, the
microarray technology has limitations including insufficient sensitivity for
quantifying lower abundant transcripts, narrow dynamic range and non-specific
17
Sequencing-based methods such as SAGE rely upon cloning and sequencing cDNA
fragments. This approach allows quantification of mRNA abundance by counting the
number of times cDNA fragments from a corresponding transcript are represented in a
given sample, assuming that cDNA fragments sequenced contain sufficient
information to identify a transcript. The currently used RNAseq is a SAGE-based
technology that allows for high-throughput analysis of transcriptomes. Its decrease in
the cost allows routinely used in research, increasing application and fast development.
In RNAseq method, complementary DNAs (cDNAs) generated from the mRNA of
interest are directly sequenced using next-generation sequencing (NGS) technologies.
The obtained reads can then be aligned to a reference genome for a whole-genome
transcriptome map.
The NGS technology enables massive parallel sequencing at a reasonable cost
[132,133]. Current NGS platforms include the Roche 454 Genome Sequencer,
Illumina's Genome Analyzer, and Applied Biosystems' SOLiD. These platforms can
analyze tens to hundreds of millions of DNA fragments simultaneously, generate
giga-bases of sequence information from a single run, and have revolutionized SAGE
18
1.5.1 RPKM measure
Measures of RNA abundance are important for many areas of biology and often
obtained from RNAseq using Illumina sequence data. These measures need to be
normalized to remove technical biases in the sequencing approach, such as the length
of the RNA species and the sequencing depth of a sample. RPKM, which is the most
frequently used measure of RNAseq data, calculates the number of reads mapped to a
particular gene region (rg) and the feature length, flg [135]. The calculation is as
RPKMg = rg x 109/flg x R, where R is the total number of reads from the sequencing run of that sample.
19
Chapter 2
YjcC, a c-di-GMP phosphodiesterase protein, regulates
the oxidative stress response and virulence of Klebsiella
20
2.1. Abstract
This study shows that the expression of yjcC, an in vivo expression (IVE) gene,
and the stress response regulatory genes soxR, soxS, and rpoS are paraquat inducible
in Klebsiella pneumoniae CG43. The deletion of rpoS or soxRS decreased yjcC
expression, implying an RpoS- or SoxRS-dependent control. After paraquat or H2O2
treatment, the deletion of yjcC reduced bacterial survival. These effects could be
complemented by introducing the ΔyjcC mutant with the YjcC-expression plasmid
pJR1. The recombinant protein containing only the YjcC-EAL domain exhibited
phosphodiesterase (PDE) activity; overexpression of yjcC has lower levels of cyclic
di-GMP. The yjcC deletion mutant also exhibited increased reactive oxygen species
(ROS) formation, oxidation damage, and oxidative stress scavenging activity. In
addition, the yjcC deletion reduced capsular polysaccharide production in the bacteria,
but increased the LD50 in mice, biofilm formation, and type 3 fimbriae major pilin
MrkA production. Finally, a comparative transcriptome analysis showed 34
upregulated and 29 downregulated genes with the increased production of YjcC. The
activated gene products include glutaredoxin I, thioredoxin, heat shock proteins,
chaperone, and MrkHI, and proteins for energy metabolism (transporters, cell surface
structure, and transcriptional regulation). In conclusion, the results of this study
21
virulence but negatively affects the biofilm formation and type 3 fimbriae expression
by altering the c-di-GMP levels after receiving oxidative stress signaling inputs.
2.2. Introduction
During infection, pathogens protect themselves from the oxidative burst of
phagocytic cells and the challenging oxidative environments within cellular and
extracellular compartments. Upon exposure to oxidative stress such as tellurite,
paraquat or hydrogen peroxide, E. coli exhibits an increase in the intracellular ROS
and the content of protein carbonyl groups [136-138]. Reactive oxygen species (ROS),
including superoxide anion (O2.-), hydrogen peroxide (H2O2), and hydroxyl radicals
(HO.), may damage DNA, proteins, and cell membranes and often lead to cell death
[117,118]. The bacterial defense mechanism includes sensing, avoiding, and
removing the ROS [123]. In general, SodA, SodB, and SodC remove superoxide,
whereas catalases (KatE and KatG) and peroxidases (AhpC and GST) remove
hydrogen peroxide [139,140]. These various stress defenses are controlled by
regulators that respond to superoxide and redox-cycling drugs (e.g., SoxRS),
hydrogen peroxide (e.g., OxyR), iron (e.g., Fur), or oxygen tension (e.g., FNR and
ArcAB) [140-143]. Diguanylate cyclases (DGCs) and phosphodiesterases (PDEs)
22
catalyzing molecular synthesis and hydrolysis, respectively [71,81]. The regulatory
roles of c-di-GMP appear in numerous bacteria in various cellular functions, including
cell surface remodeling [144], cellulose synthesis [111], virulence [145], motility
[107], and biofilm formation [146-148]. E. coli YfgF, which exhibits PDE activity,
regulates not only surface cell remodeling but also the oxidative stress response by
modulating c-di-GMP levels [11]. The disruption of Salmonella enteric Var.
typhimurium cdgR, which encodes a PDE protein, also decreases bacterial resistance
to hydrogen peroxide and accelerates death by macrophages [149].
Klebsiella pneumoniae pyogenic liver abscess isolates often carry heavy capsular
polysaccharides (CPS) to avoid phagocytosis or death by serum factors [33,150]. This
thick and viscous structure also helps regulate the bacterial colonization and biofilm
formation at the infection site [151]. Several regulators, such as RcsB, RmpA,
RmpA2, KvhR, KvgA, and KvhA, help control the CPS biosynthesis by regulating
the cps transcriptions in K. pneumoniae [39,152]. An increase in CPS synthesis
protects K. pneumoniae from oxidative stress [32,153,154]. However, whether the
modulation of c-di-GMP affects CPS synthesis remains unclear.
The expression of yjcC, an IVE gene isolated from the liver abscess isolate K.
pneumoniae CG43, is inducible in the presence of 10 M paraquat [115]. Sequence
23
CSS motif at the N-terminal region, whereas the C-terminal contains a conserved
EAL domain of the PDE enzyme [155]. In addition, the encoding gene yjcC is
cluster-located with soxRS genes, suggesting that it plays a role in the oxidative stress
response. This study investigates whether YjcC plays a role in oxidative stress
defenses and if YjcC uses PDE activity to execute its regulation.
2.3. Results
2.3.1The YjcC expression is paraquat inducible, and SoxRS and RpoS dependent
To confirm the previously reported paraquat-induced expression phenotype [115],
the IVE DNA containing the 5’ non-coding region and part of the coding sequence of
yjcC was isolated from K. pneumoniae CG43S3 and cloned in front of the
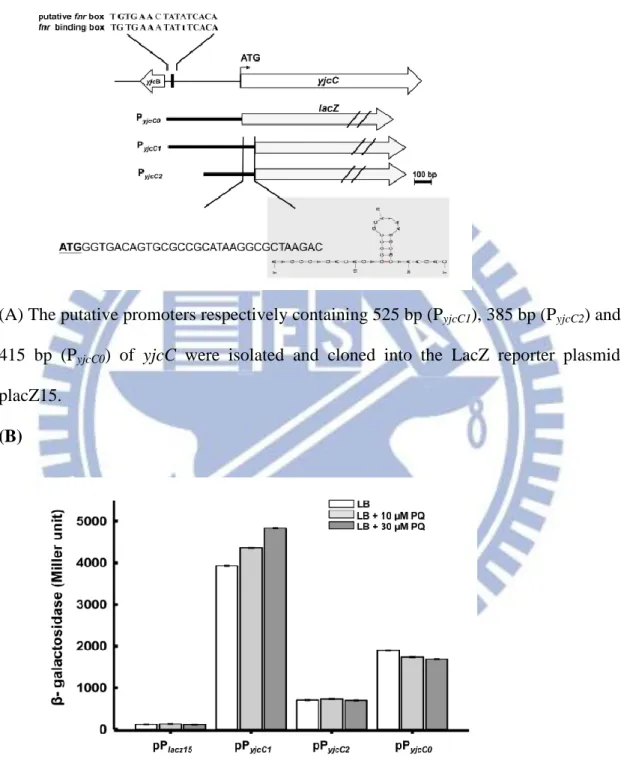
promoterless lacZ gene of pLacZ15 [152]. The resulting plasmid was called pPyjcC1.
The sequence analysis of PyjcC1 shows a conserved Fnr box TGTGA-N6-TCACA [156]
centered approximately 400-bp upstream of the yjcC start codon. This process also
generated recombinant plasmids pPyjcC2 and pPyjcC0 carrying truncated forms of PyjcC1.
These plasmids respectively removed the putative Fnr box and the small stem-loop
sequence of the 33-bp coding region shown in Fig.2.1 (A). As Fig. 2.1 (B) shows, the
bacteria containing pPyjcC1 exhibited the highest level of -galactosidase activity,
24
not PyjcC2 nor PyjcC0, increased after adding 10 μM of paraquat to the culture medium.
This paraquat-induced characteristic also appeared when the concentration increased
to 30 μM, further enhancing the activity of PyjcC1.
As Fig. 2.1 (C) shows, the addition 30 μM paraquat to the bacterial culture
significantly increased the yjcC mRNA level. Compared to the expression of the
well-characterized stress response regulators SoxS, SoxR, RpoS, and Fnr, the yjcC
gene expression was more responsive to paraquat than to hydrogen peroxide exposure.
This study also investigates whether yjcC is subjected to regulation by SoxRS or
RpoS. As Fig.2.1 (D) shows, the deletion of soxR, soxS, or rpoS reduces the yjcC
expression, implying that SoxRS and RpoS play a positive role in yjcC expression.
2.3.2 YjcC plays a positive role in the oxidative stress response
Paraquat is a superoxide anion generator. Thus, the paraquat-inducible expression
suggests that YjcC plays a role in the oxidative stress response. To investigate this
possibility, an yjcC deletion mutant was generated through an allelic exchange
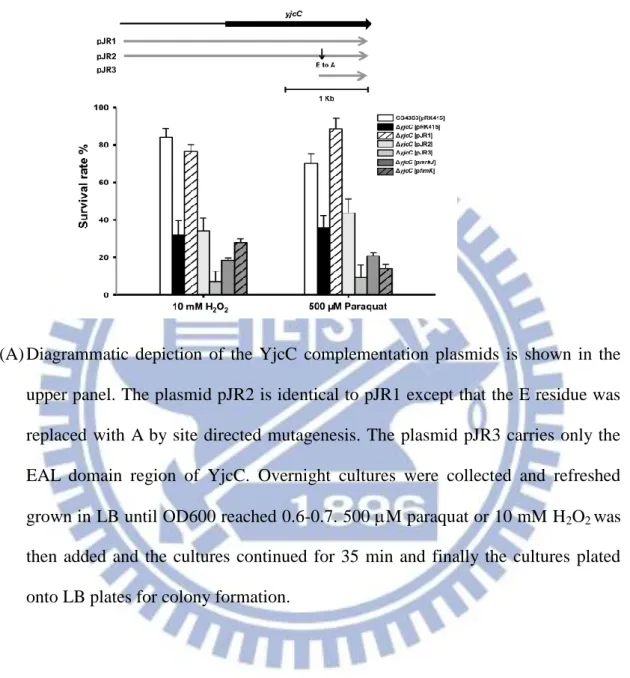
strategy. As Fig. 2.2 (A) shows, the yjcC deletion mutant was more sensitive to
paraquat and hydrogen peroxide when compared to the wild type bacteria K.
pneumoniae CG43S3. The deletion effect could be complemented by transforming the
25
which expresses the mutant form of YjcC with the conserved E residue of the EAL
domain replaced by A or pJR3 (which carries the coding region of the YjcC EAL
domain), had no complementation effect. Neither of the two EAL-domain protein
encoding plasmids pmrkJ and pfimK, which carry PDE activity, could complement the
yjcC deletion effect. These results suggest that the stress response is YjcC dependent
and both the N-terminal signaling receiving region and the EAL domain of YjcC are
required and specific for an oxidative stress response.
To determine if the YjcC-EAL domain exhibits PDE activity, the recombinant
expression plasmid containing the DNA coding for the EAL domain of YjcC or the
AAL coding region of pJR2 was constructed and overexpresed in E. coli, and the
recombinant proteins were purified. Figure 2.2 (B) shows that the purified EAL
domain protein exhibits PDE activity towards pNpp. This activity is lower than the
level of the recombinant MrkJ [157], but considerably higher than the activity of the
recombinant protein AALyjcC. As Fig.2.2 (C) shows, the c-di-GMP level of
CG43S3ΔyjcC[pJR1] was significantly lower than those of CG43S3[pRK415],
CG43S3ΔyjcC[pJR2], or CG43S3ΔyjcC[pJR3]. This suggests that YjcC in vivo
functions as a PDE enzyme capable of reducing the intracellular c-di-GMP levels. The
deletion of yjcC gene from CG43S3 increased the c-di-GMP amounts and the
26
30 µM paraquat Fig. 2.2(D). This also suggests that YjcC is able to degrade c-di-GMP
and the catalytic activity could be enhanced by oxidative stress.
2.3.3 Deletion of yjcC places bacteria in an oxidative stress state
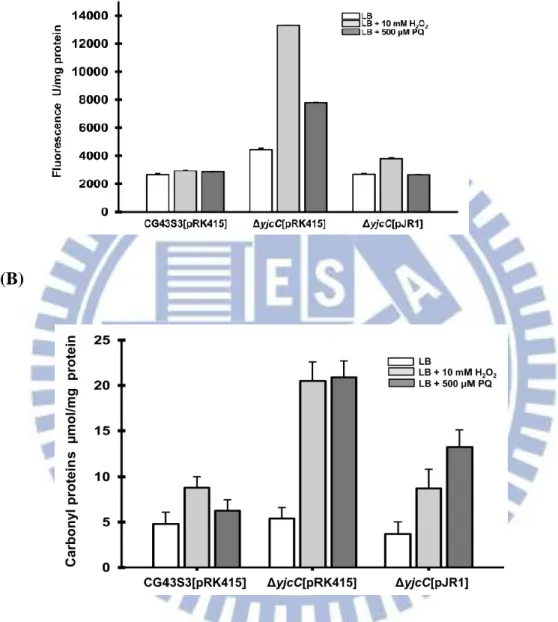
As Figs. 2.3 (A) and (B) show, the deletion of yjcC after treatment of H2O2 or
paraquat significantly raised the levels of the fluorescent probe H2DCFDA (used to
monitor the formation of ROS) and carbonyl proteins. The introduction of pJR1 into
CG43S3ΔyjcC mutant appeared to reduce the levels of ROS and the carbonyl proteins,
showing that YjcC is involved in the removal of ROS or damaged molecules. Thus,
this study also investigates the anti-oxidant activity of YjcC. As Fig. 3C shows, the
deletion of yjcC reduced the oxidant scavenging activity, as assessed by the
absorbance change at 517 nm for the decolorization degree of the purple color,
supporting the possibility that YjcC modulates anti-oxidant activity in a certain
manner. Numerous studies have shown that Fur and RpoS affect and regulate
numerous SODs and catalases [139,158-161]. Figure 2.3 (D) shows that zymogel
analysis and total activity measurement exhibit significant changes in the SOD or
catalase activity after the deletion of fur or rpoS. However, the deletion of yjcC has no
apparent influence on SOD or catalase activity, suggesting that the YjcC-dependent
27
2.3.4 YjcC plays a regulatory role in the virulence, CPS production, biofilm
formation, and type 3 fimbriae expression
YjcC, previously identified as an IVE gene product, is likely involved in infection
[115]. To investigate whether YjcC is a virulence factor for the bacteria to establish
infection, a mouse peritonitis model was employed. As Table 1 shows, the LD50 to
Balb/c mice increased approximately 10-fold after yjcC deletion; introducing ΔyjcC
with pJR1, but not pJR2 or pJR3, could restore the LD50. This indicates that YjcC
expression at a certain stage is required for mouse infection. It is interesting to note
that the ΔyjcC colony is smaller and less mucoid, as determined by a string test [39],
than its parental strain on LB agar plate. Therefore, sedimentation analysis and
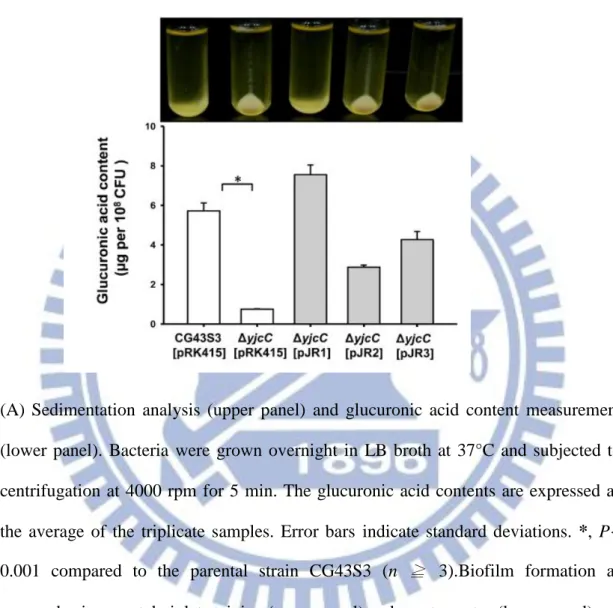
glucuronic acid content measurement are carried out to determine the CPS production.
As Fig. 2.4 (A) shows the deletion of yjcC reduces CPS production. The CPS
deficient phenotype can be fully complemented with the transformation of pJR1 into
the ΔyjcC mutant. However, transforming the mutant with pJR2 or pJR3 partially
restores glucuronic acid production.
The second messenger c-di-GMP plays an important role in bacterial biofilm
formation [14-16]. Figure 2.4 (B) shows that the biofilm formation activity of ΔyjcC
appears to increase compared to the parental strain, whereas that transformed with
28
c-di-GMP (Fig. 2.2 C), which indicates that the YjcC expression of pJR1 significantly
reduces the c-di-GMP level, thus reducing biofilm forming activity. Moreover, type 3
fimbriae is a major determinant of biofilm formation in K. pneumoniae [64].
Therefore, this study also investigates the deletion effect on type 3 fimbriae
expression. As Fig. 2.4 (C) shows, the western blot hybridization with anti-MrkA
antibody shows that the yjcC deletion also increased the major pilin MrkA production
of type 3 fimbriae.
2.3.5 Effects of YjcC overexpression assessed using a transcriptome study
This study uses comparative transcriptome analysis between CG43S3[pRK415] and
CG43S3[pJR1] to gain further insights into how YjcC executes its regulation.
Analysis of the genome annotation of liver abscess isolate K. pneumoniae
NTHU-K2044 [112] shows that the increased expression of yjcC significantly
enhances the expression of 34 genes. As Table 2.2 shows, the YjcC-activated genes
can be categorized into 12 functional groups. These include the oxidative stress
response genes grxA, ybbN, dinI, priB, and stpA, which are involved in anti-oxidation
[162,163] or DNA repair [163], the heat shock chaperone protein encoding genes
ibpB, ibpA, htpG, and dnaK, which are generally induced in stress conditions [164],
29
aggregation and help maintain proton motive force (PMF) to counteract stress
conditions [165,166]. Increasing the expression of yjcC also enhanced the expression
of PilZ domain protein MrkH and the LuxR-type transcription factor MrkI.
Conversely, 29 genes whose expressions were significantly repressed by the increase
of YjcC expression include fumB, which is regulated by fnr under limited oxygen and
anaerobic conditions [124]. Other YjcC negatively affected genes are metabolite
transporter genes and genes coding for permease and energy metabolism involved in
the synthesis of amino acids (Table 2.3).
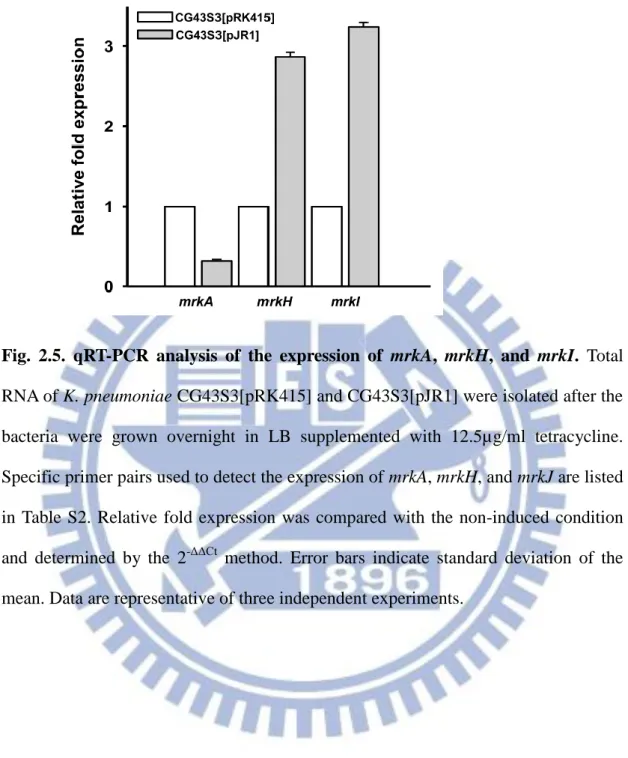
As assessed by qRT-PCR analysis, Fig. 2.5 shows that the mRNA level of mrkH
and mrkI, respectively increased 2.87- and 3.24-fold in CG43S3[pJR1] compared to
that of CG43S3[pRK415]. In contrast, the mrkA transcript levels dropped to
approximately 1/3 of that of CG43S3[pRK415].
2.4. Discussion
In E. coli, all the described genes inducible by paraquat are a part of the SoxRS
regulon [167-169]. The expression of yjcC is RpoS-dependent in E. coli [170].
Consistent with this finding, the yjcC expression in K. pneumoniae is affected by
RpoS and by SoxRS at the transcriptional level. No SoxRS or RpoS binding box
appears within the putative promoter sequence, suggesting the possibility of an
30
and anaerobic growth at the transcriptional level [171]. The conserved fnr binding box
present in the upstream non-coding region of yjcC implies an FNR dependent control
of YjcC expression. Thus, the yjcC regulation by FNR likely occurs in poorly
oxygenated environments.
The K. pneumoniae NTUH K2044 genome contains 27 genes encoding
GGDEF-, EAL-, and GGDEF-EAL-domain proteins of potential DGC and PDE
enzyme activity [172]. The yjcC encoding gene is also identified as member of the
protein family. The family regulation specificity is determined by the sensory domain
of the DGC or PDE proteins. Figure 2.2 (A) shows that the PDE expression plasmids
pfimK and pmrkJ, which contain the respective coding region with putative promoter,
failed to complement yjcC deletion. This may be due to fimK and mrkJ genes are not
induced in comparison to yjcC in the presence of paraquat. There is also the
possibility that the N-terminal peptide of approximately 300 aa of YjcC plays a role in
the oxidative stress response besides signal sensing. Various sensory domains can
bind to small molecular signals, and through this connection, modulate the levels of
c-di-GMP [173,174]. Although the signal for the CSS-motif remains unknown, we
speculate that YjcC-mediated signaling sensing may occur in the periplasmic space
because of the signal peptide and the transmembrane domain at the N-terminal region.
31
amount of c-di-GMP in CG43S3ΔyjcC[pJR3] is approximately the same as that in
CG43S3ΔyjcC[pRK415] and CG43S3ΔyjcC[pJR2], suggesting that the EAL domain
has extremely low levels of PDE activity. The N terminal region of YjcC likely
requires receiving some signal from outside, and the interaction of the sensory domain
with signaling molecules activates the PDE activity. This is supported by Fig. 2.2 (D)
showing that the deletion of yjcC gene from CG43S3 raised the c-di-GMP levels and
the influence was much more apparent after the bacteria exposure to 30 µM of
paraquat.
The deletion of yjcC decreases the CPS production and the virulence attenuation,
indicating that YjcC is required to avoid the damage of oxidative stress. In bacteria,
superoxide dismutase and catalase are common antioxidant enzymes that scavenge
ROS from oxidative stress. The redox proteins (including GrxA and YbbN) required
for maintaining redox status in bacteria are also protect bacteria from oxidative stress
[175,176]. The deletion of yjcC has no apparent effect on the SOD and catalase
activity, but appears to increase the transcription of grxA and ybbN. This suggests that
YjcC is involved in regulating the redox levels in bacteria after oxidative stress. The
chaperone protein ClpB stabilizes protein and suppresses the protein aggregation
induced by heat or other stresses [165,177]. The phage shock proteins, PspB and PspC,
32
interaction, maintaining the proton-motive force under extracytoplasmic stress
conditions [166]. Transcriptome analysis shows that an increase in YjcC induces the
expression of several heat shock proteins and chaperones, suggesting that YjcC is
involved in regulating anti-stress responses. The fumarase gene fumB, which is
expressed under anaerobic cell growth conditions, is regulated by Fnr and ArcA
[178,179]. The result of yjcC expression reduced the fumB transcripts implies that
YjcC is a component of the Fnr and ArcA regulatory pathway.
In K. pneumoniae, the second messenger c-di-GMP activates type 3 fimbriae
expression through MrkHI activation [146]. However, the increased synthesis of
mrkH and mrkI transcripts by the overexpression of yjcC remains unclear. We have
found that mrkHI expression is barely detected under LB culture (unpublished
observation). Under oxidative stress pressure, the N-terminal region of YjcC may turn
on the expression of MrkHI. Thus, the N-terminal region likely plays a determinant
role in YjcC-dependent regulation. E. coli YdeH has c-di-GMP cyclase activity [180].
The transcriptome analysis as shown in Table S3 revealed that the transcript levels of
mrkA, mrkH, and mrkI in CG43[pRK415-ydeH], with c-di-GMP level of 23.1 fmole/
mg-1 (data not shown), significantly increased compared to those of CG43S3[pJR1].
This also indicates that mrkHI expression is c-di-GMP level-dependent and the
33
Overall, these results indicate that the YjcC-mediated regulatory system is
considerably more complex than expected. During infection, the transition from
aerobic to microaerobic conditions or the transition from a microaerobic to oxidative
stress environment, YjcC may be activated through sensory regulatory systems on the
N-terminal region. Thereafter, YjcC modulates the levels of c-di-GMP to affect the
expression of the downstream regulatory pathways. In conclusion, YjcC regulates the
oxidative stress response, mouse virulence, CPS synthesis, biofilm formation, and
type 3 fimbriae expression. This most likely occurs through the adjustment of
34
The tested bacterial strains CG43S3, CG43S3ΔyjcC,CG43S3ΔyjcC[pJR1] and CG43S3ΔyjcC[pJR2] were cultured in LB medium at 37°C for overnight.
Five mice of a group were injected intraperitoneally with bacteria resuspended in 0.2 ml of saline in 10-fold steps graded doses. The LD50, based on the number of survivors after one week, were calculated by the method of Reed and Muench [215]and expressed as CFU.
Table 2.1. YjcC effect on the mouse virulence.
Strain LD50
CG43S3 1x104
CG43S3ΔyjcC 7.8 x105
CG43S3ΔyjcC [pJR1] 3.3 x104
35
Table 2.2 Significantly upregulated genes by yjcC overexpression
Proposed function Gene name Folda expression ORFb ID Oxidative response
/repair/sos responsive
Glutaredoxin I grxA 2.6 KP1_1843
Thioredoxin ybbN 2.4 KP1_1349
DNA damage inducible protein I dinI 2.1 KP1_2061
Heat shock response/chaperones/protein modification
Heat shock protein ibpB 5.9 KP1_5467
Heat shock protein ibpA 5.6 KP1_2622
Heat shock protein 90 htpG 2.9 KP1_1331
Heat shock protein 70 dnaK 3.5 KP1_0835
Protein disaggregation chaperon clpB 3.2 KP1_4170
Phage shock protein B pspB 2.1 KP1_2344
Ci-di GMP metabolite
EAL containing protein yjcC 6.3 KP1_0324
PilZ domain protein mrkH 3.2 KP1_4551
Energy/intermediary metabolism
Carbamoyl phosphate synthase carA 3.2 KP1_0853
Ascorbate specific PTS family enzyme 3.1 KP1_2793
D-arabinitol dehydrogenase 2.9 KP1_3760
Allulose-6-phosphate 3-epimerase 2.3 KP1_2791
Xylulokinase 2 KP1_3759
Transporter
ABC transporter ATP-binding protein 2.5 KP1_5267
Putative transport protein/kinase ydjN 2 KP1_2272
Amino acid biosynthesis
Sulfate adenylate transferase subunit 2 cysD 2.4 KP1_4384
Nucleotide biosynthesis and metabolism
Methionine sulfoxide reductase A msrA 2.5 KP1_0489
DNA replication/recombination/repair
Primosomal replication protein N priB 2.1 KP1_0469
DNA binding protein, nucleoid associated stpA 2.6 KP1_4260
Cell surface structures
Prepillin peptidase dependent protein 2.6 KP1_0311
Two component system connector ycgZ 2.6 KP1_1728
Pullulanase-specific type II secretion system outer membrane lipoprotein
pspD 2.3 KP1_0998
Regulators
LysR family transcriptional regulator 2.8 KP1_4352
Transcriptional activator nhaR 2.1 KP1_0838
LuxR family regulatory protein mrkI 2.6 KP1_4552
Hemolysin modulating protein hha 2.6 KP1_1317
Transcriptional antiterminator of glycerol uptake operon
2.1 KP1_1112
DNA binding transcriptional activator pspC 2.1 KP1_2344
Hypothetical proteins
ParB family protein HP 3.3 KP1_2152
36
Table2 and 3. Significantly upregulated and downregulated genes by YjcC
overexpression. The selected genes show more than 2 log2 fold change (in absolute value) in the abundance of transcript between K. pneumoniae CG43S3[pRK415] and
Table 2.3. Significantly downregulated genes by yjcC overexpression
Proposed function Gene name Folda
expression
ORFb ID
Anaerobic response protein
Anaerobic class I fumarate hydratase fumB -5.7 KP1_2562
Transporter
Alanine/serine/glycine transport protein -6.8 KP1_2505
ABC transport system ATP binding protein -4.5 KP1_3173
PTS transporter subunits II ABC -5.0 KP1_3804
Methylgalactoside transporter inner membrane component mglC -4.8 KP1_0277
Methylgalactoside transporter system Substrate-binding component
mglB -4.2 KP1_3815
Putative ABC transport system component -4.6 KP1_3175
Molybdate ABC transporter system -4.4 KP1_3995
Maltose/maltodextrin transporter ATP binding protein malK -4.2 KP1_0276
Sugar ABC transport system permease component -4.1 KP1_1424
Putative ABC transporter mocB -3.7 KP1_1423
Putative rhizopine uptake ABC transporter proY -3.7 KP1_1422
Permease
Putative PTS permease -6.5 KP1_0760
Putative amino acid permease -4.3 KP1_1204
Amino acid biosynthesis
Arginine succinyltransferase -4.5 KP1_2499
Acetylornithine transaminase -5.0 KP1_2498
Putative glutamine synthetase -3.3 KP1_2006
Energy/intermediary metabolism Succinate antiporter -6.4 KP1_2563 Succinylarginine dihydrolase -5.3 KP1_2502 Glucosamine-fructose-6-phosphate aminotransferase -6.0 KP1_0764
NADH: flavin oxidoreductase -5.8 KP1_2565
Phospho-beta glucosidase -5.2 KP1_3803
Acety-CoA synthetase asc -4.3 KP1_0342
Succinylglutamate desuccinylase astE -4.0 KP1_2256
Phosphomannosemutase manB -3.4 KP1_3702
Putative monooxygenase subunit -3.1 KP1_1996
Putative acid phosphotase -3.1 KP1_3725
Regulators
DNA binding transcriptional repressor lldR -4.5 KP1_5297
Cell surface structures
37
CG43S3[pJR1]. a. Log2 fold change represents the log2 ratio of mRNA transcript levels of CG43S3[pJR1] to CG43S3[pRK415]. b. Open reading frame (ORF) ID is as annotated from K. pneumoniae NTUH K2044.
38
Fig. 2.1. The yjcC is paraquat inducible, and SoxRS and RpoS dependent. (A)
(A) The putative promoters respectively containing 525 bp (PyjcC1), 385 bp (PyjcC2) and
415 bp (PyjcC0) of yjcC were isolated and cloned into the LacZ reporter plasmid
placZ15.
(B)
(B) The recombinant plasmids placz15, pPyjcC1, pPyjcC2 and pPyjcC0 were then
transformed to K. pneumoniae CG43Z01 and the -galactosidase activities of the transformants grown to log-phase in LB broth were determined. The results are shown as an average of triplicate samples. Error bars indicate standard deviations.



![Table 3.1 and 3.2 Up-regulated genes and down-regulated genes from the comparison of CG43S3[pRK415-ydeH] and CG43S3[pRK415]](https://thumb-ap.123doks.com/thumbv2/9libinfo/8213388.170152/67.892.135.809.212.1079/table-regulated-genes-regulated-genes-comparison-cg-ydeh.webp)