行政院國家科學委員會專題研究計畫 成果報告
油菜固醇對綠豆上胚軸延長及低溫反應的蛋白質體與代謝
質體研究
研究成果報告(精簡版)
計 畫 類 別 : 個別型 計 畫 編 號 : NSC 95-2311-B-002-007- 執 行 期 間 : 95 年 08 月 01 日至 96 年 11 月 30 日 執 行 單 位 : 國立臺灣大學植物科學研究所 計 畫 主 持 人 : 陳益明 共 同 主 持 人 : 陳淑玲 計畫參與人員: 助理教授:陳淑玲 副教授:阮雪芬 博士班研究生:黃斌 報 告 附 件 : 出席國際會議研究心得報告及發表論文 處 理 方 式 : 本計畫可公開查詢中 華 民 國 97 年 02 月 14 日
油菜固醇對綠豆上胚軸延長及低溫
反 應 的 蛋 白 質 體 與 代 謝 質 體 研 究
期末報告
主 持 人:陳益明
計畫編號:NSC95-2311-B-002-007
執行期間:95 年 8 月 1 日至 96 年 11 月 30 日
Elucidating the Growth Regulation of Brassinosteroids in Mung
Bean Epicotyls using a Proteomics Approach
Bin Huanga, Kuo-Chin Nib, Shu-Ling Chenc, Hsueh-Fen Juanb,d,e*, Yih-Ming Chena,d*
aInstitute of Plant Biology, National Taiwan University, Taipei, Taiwan
bInstitute of Molecular and Cellular Biology, National Taiwan University, Taipei,
Taiwan
cDepartment Biotechnology, Ming Chuan University, Taoyuan, Taiwan dDepartment of Life Science, National Taiwan University, Taipei, Taiwan
eCenter for Systems Biology and Bioinformatics, National Taiwan University, Taipei,
Taiwan
*
Corresponding authors:
Professor Hsueh-Fen Juan, Department of Life Science & Institute of Molecular and Cellular Biology, National Taiwan University, Taipei, Taiwan, No 1, Sec. 4, Roosevelt Road, Taipei, 106 Taiwan
E-mail: yukijuan@ntu.edu.tw Tel: +886-2-33664536
Fax: +886-2-23673374
Professor Yih-Ming Chen, Department of Life Science & Institute of Plant Biology, National Taiwan University, Taipei, Taiwan, No 1, Sec. 4, Roosevelt Road, Taipei, 106 Taiwan
E-mail: yihmingc@ntu.edu.tw Tel: +886-2-33662524
Fax: +886-2-83695080
Key words: Proteomics; Brassinosteroid; Mung bean; Epicotyl
Abbreviations: BR, brassinosteroid; EBL, 24-epibrassinolide; PAG,
Abstract
Brassinosteroids (BRs), structurally similar to animal steroid hormones, are ubiquitously distributed through out the plant kingdom with significant growth-promoting activity. Even though the BR-regulated transcriptome has been revealed, the proteomics data are still rare. Mung bean seedlings treated with 24-epibrassinolide (EBL), a kind of BRs, exhibited significant epicotyl elongation. Here, we present a proteomics study of EBL-treated mung bean epicotyls. Many researches showed that de novo sequencing is very useful to identify the proteins in non-model organisms. We successfully used de novo sequencing to identify 12 differentially expressed proteins in mung bean treated with or without EBL. These proteins are mostly involved in cell growth, anti-stresses, and cell metabolism including respiration and photosynthesis. In the physiological assay, the retarded growth caused by inhibitors of methionine synthase and enolase could be recovered by exogenous treatment of EBL. Based on the identified proteins, we proposed a possible mechanism of BR-promoted cell elongation. This study confirms that proteomic analysis is a potent tool for analyzing the protein expression related to the effect of steroid hormones on epicotyl growth. Our approach opens a way of defining hormonal targets for plant growth. This not only elucidates a molecular mechanism of BR regulation but may also be extended for plant breeding.
1 Introduction
Brassinosteroids (BRs) are growth-promoting natural products that comprise a class of over 40 poly-hydroxylated sterol derivatives, and are found at low levels in pollen, seeds, and young vegetative tissues [1]. With combinations of HPLC, GC-MS techniques and recent achievements in genetic research (such as mutants with deficient BR biosynthesis and signaling pathways from Arabidopsis, pea, tomato and rice), the biosynthetic pathway including early and late C6-oxidation and signaling components , such as BRI1, BAK 1, BZR1 and BES1, have been identified [2-5]. In addition to the growth-promoting activity, the involvements of BR with seed germination, vascular development, flowering, senescence, and stress tolerance are also reported [6]. These studies confirmed predictions of plant physiologists that BR is essential for plant growth and must be considered along with phytohormones such as auxin, cytokinins, gibberellins, abscisic acid, and ethylene in any model of plant development [7]. These form consensus among scientists to regard BRs as the sixth class of phytohormone.
Mung bean (Vigna radiata L.), an important tropical crop, has green skin and is also called Green Bean in China. It has been the subject of discussion for BR-induced cell elongation ever since BR was discovered [8,9]. Different from model plants such as Arabidopsis and rice, epicotyl is a specific tissue found in the legumes that has been used in molecular study for the mechanisms of cell elongation [10,11]. The downstream components of BR on promoting cell growth have been discussed at the transcriptome level [12,13]; however, little proteomics-based research has been done on BR-promoted cell growth. Due to the post-transcriptional modification and alternative splicing, the levels of mRNA do not necessarily predict the levels of corresponding proteins in physiological responses. Therefore, the proteomics study is considered worthy to explicate the molecular mechanisms of elongation in mung bean
epicotyls regulated by BRs.
Proteomics is the systematic analysis of the proteome, the totality of proteins expressed by a genome [14]. Two-dimensional gel electrophoresis coupled with protein identification by mass spectrometry founded proteomics platform upon differential expression and chemical treatment studies in plants. Conventional techniques for MS-based protein identification rely on exact matching of masses calculated from sequences from database entries; consequently, these methods fail to identify, or would misidentify proteins if the sequence is not present in these databases [15]. Since existing databases lack sufficient amino acid sequence homology with protein for non-model organisms, the identification of short peptides from mung bean is arduous. Thus, a database search strategy that encompasses both identity and homology can provide stronger evidence than a single search alone [16]. Using de novo sequencing is very useful to identify the proteins in non-model organisms. PEAKS is a powerful software for de novo sequencing by tandem mass spectrometry that has been cited about 60 times. With de novo sequencing, a reconstruction of the peptide sequence is done without a protein database, and is then followed by query against the NCBInr and Swiss-Prot databases using BLAST or Mass spectrometry-driven BLAST (MS BLAST) tools. MS BLAST is a database search protocol used for identifying unknown proteins by sequencing similarity to homologous proteins available in the database [17,18]. In this study, we successfully identified the BR-regulated proteins in mung bean. Furthermore, we infer a possible molecular mechanism in BR-regulated growth effects.
2 Materials and methods
2.1 Plant materials, growth conditions and inhibitor treatments
To allow the first true leaf to fully open, mung bean (Vigna radiata L., V2937) and soybean (Glycine max cv. Kaohsiung No.8) seedlings were germinated on moist vermiculite deprived of nutrients in a growth chamber for five days and ten days respectively. The growth conditions were set at 28 ℃ with 70 % relative humidity, 16 hr light, 8 hr dark, and a light intensity of 200 μmolm-2s-1. Each pot contained ten unique 5-day-old or 10-day-old seedlings, and underwent the subsequent treatments in this study. The stocks of chemicals were from Sigma (MO, USA): 24-epibrassinolide (EBL, 3 mM/DMSO), DL-propargylglycine (PAG, 10 mM/NaOH) and potassium fluoride (KF, 10 mM/H2O). Each pot containing ten unique seedlings was treated with
serial dilutions with chemicals or equal amount of solvent onto shoot meristem and allowed to grow for one day. The optimal concentration of chemicals applied was determined by the epicotyl elongation rate, which was calculated by dividing increased length by original length. The inhibitor-treated and the control plants were allowed to grow for two days instead of one to allow for greater differences between the two treatments. Different light intensity of 200 μmolm-2s-1 (light), 80 μmolm-2s-1 (40 % light) and full darkness were designated to investigate the interdependency between light and BR.
2.2 Light microscopy
A 2 mm section cut 1 cm below the apical meristem was fixed with 2.5 % (v/v) glutaraldehyde in 0.1 M phosphate buffer (pH 7.0) at room temperature for 2 hr and was post-fixed with 1 % (w/v) osmium tetraoxide at room temperature or overnight at 4 ℃. The specimens were dehydrated in a grade acetone series and were embedded in Spurr’s resin. Semi-thin sections with 1 μ m thickness were prepared by
Ultramicrotome (Reichert Jung Ultracut E, Germany) and stained with toluidine blue.
2.3 Protein extraction
Mung bean epicotyls from EBL or Control treatments were separately grounded with liquid nitrogen and suspended with an extraction buffer: 50 mM Tris-HCl (pH 8.0), 3 mM phenylmethylsulfonyl fluoride (PMSF), 1 % (v/v) 2-mercaptoethanol, 0.2 % (v/v) Triton X100, 10 % (w/v) sucrose and 10 mM ascorbic acid with 1:5 tissue/buffer ratio. After centrifuge at 12000g for 10 min, the supernatants were precipitated with acetone containing 10 % (w/v) trichloroacetic acid (TCA) and 0.3 % (w/v) dithiothreitol (DTT) at -20 ℃ overnight.
2.4 2-DE analysis
The dried protein pellets were dissolved in the sample buffer: 7 M urea, 2 M thiourea, 4 % (w/v) CHAPS, 60 mM DTT and 2 % (v/v) IPG buffer. The epicotyl proteins (300 µg) were mixed with DeStreak Rehydration solution and soaked into 18 cm DryStrip (Amersham Pharmacia Biotech) for rehydration on the Ettan IPGphor system. The voltage was set to 32 kVhr for DryStrip pH 4-7 and 61.5 kVhr for DryStrip pH 4.5-5.5. After IEF analysis, the stripped gels were subjected to reduction with 1 % (w/v) DTT in an equilibration buffer: 2 % (w/v) SDS, 50 mM Tris-HCl (pH 8.8), 6 M urea and 30 % (v/v) glycerol, and followed by alkylation with 2.5 % (w/v) iodoacetamide (IAA) in the same buffer. The equilibrated gels were attached with 0.5 % agarose to the top of a vertical 12.5% SDS-PAGE system modified from Laemmli [19].
2.5 Image analysis and In-gel protein digestion
The 2-D electrophoresis gels were stained with SYPRO-Ruby (Molecular Probes, OR, USA) and digitally scanned using a Typhoon 9400 fluorescence image scanner
(Amersham Bioscience, NJ, USA). Protein spots were automatically detected and analyzed using ImageMaster software (Amersham Bioscience). Spot intensity levels were normalized between gels as a proportion to the total protein intensity detected for the entire gel. The experiments were repeated three times; only the protein spots that corresponded to be regulated by EBL were excised and subjected to the in-gel digestion process according to the guidelines given in the In-Gel Tryptic Digestion Kit (Pierce, IL, USA) and research articles [20].
2.6 Mass Spectrometry Analysis
The lyophilized samples were re-suspended in 0.1 % trifluoroacetic acid (TFA). The matrix, alpha-cyano-4-hydroxycinnamic acid (CHCA), was premixed in 30% acetonitrile (ACN)/0.1% TFA solution (w/v). The sample and CHCA mixture were loaded onto the MALDI plate (PerSeptive Biosystems, CA, USA) via the dried droplet method [21]. MALDI-quadrupole (Q)-TOF-MS was performed on a Voyager-DE PRO (PerSeptive Biosystems) operating in reflector mode. The instrument was equipped with a 337-nm nitrogen laser source operating at a frequency of 3-20 Hz. The spectra were recorded in reflector mode at an acceleration voltage of 20 kV, 70 % grid voltage, 0 % guide wire voltage, 100 ns delay and a low mass gate of 500 Da. Fifty laser shots were averaged for a typical spectrum. The mass calibration of the spectra was obtained by using mixtures of three reference peptides (human angiotensin I, ACTH clip 1-17 and ACTH clip 18-39) covering the m/z 500-3500 range. The most intense ions in the TOF-MS spectrum were selected for optimized MS/MS analysis.
2.7 Protein Identification
program (http://www.matrixscience.com/). Mass tolerance was set at 50 ppm for the masses of peptide precursors and at 0.15 or 0.8 Da for the masses of fragment ions. No search parameters were imposed to limit species specificity, protein pI or molecular weights [15,16]. We used de novo sequencing software, PEAKS, to confirm the protein identification [22-24]. PEAKS is a powerful software for de novo sequencing by tandem mass spectrometry and has been cited about 60 times. With de
no sequencing, a reconstruction of the peptide sequence is done without a protein
database, followed by query against the NCBInr and Swiss-Prot databases using BLAST or MS BLAST tools [17,18].
2.8 Statistics
3 Results and discussions
3.1 Exogenous treatment of BR elongated mung bean epicotyls
Brassinolide (BL), 24-epibrassinlide (EBL) and 28-homobrassinolide are the three biologically active BRs that have been widely used [7]. In this study, ten micromoles of EBL were determined as the optimal concentration used because it showed the most significant elongation in epicotyls within one day (Fig. 1A and 1D). Regarding to the chemical treatments of epicotyl meristem, the equal elongation of three-divided sections of whole epicotyl showed a possible transportation of BR on mung bean epicotyl (Fig. 1B and 1E), even though the BR does not undergo long-distance transport in pea have been proposed [25]. Soybean, the most investigated legume, has been used for molecular investigation on epicotyl elongation [10,11]. In this study, a similar EBL-response was also observed in EBL-treated soybean epicotyl (Fig. 1C), while its elongation rate (Control: EBL = 2.13) was little lower than mung bean (Control: EBL = 3.16, Fig. 1F). Therefore, mung bean epicotyl was selected to conduct proteomic regulations for BR-promoted cell growth. As for the BR-induced cellular variations, pith cells showed more expanded after EBL treatment in both cross-section and longisection under light microscopy observation (Fig. 2).
3.2 Differential expression of proteins
First, we monitored the expression profile of mung bean epicotyls in response to treatment of EBL. Protein analysis of the epicotyls treated with or without EBL for 24 hours was done using 2-D gel electrophoresis. Gel electrophoresis was first performed with pH 3-10 nonlinear strips and 12.5% SDS polyacylamide gel. The 2-DE gels were digitized and analyzed simultaneously with ImageMaster software. In the 2-DE map with pH 3-10 IPG DryStrip, a total of 572 proteins were detected on SYPRO-Ruby stained gel range. The 2-DE separation efficiency with pH 4-7 and 4.5-5.5 IPG
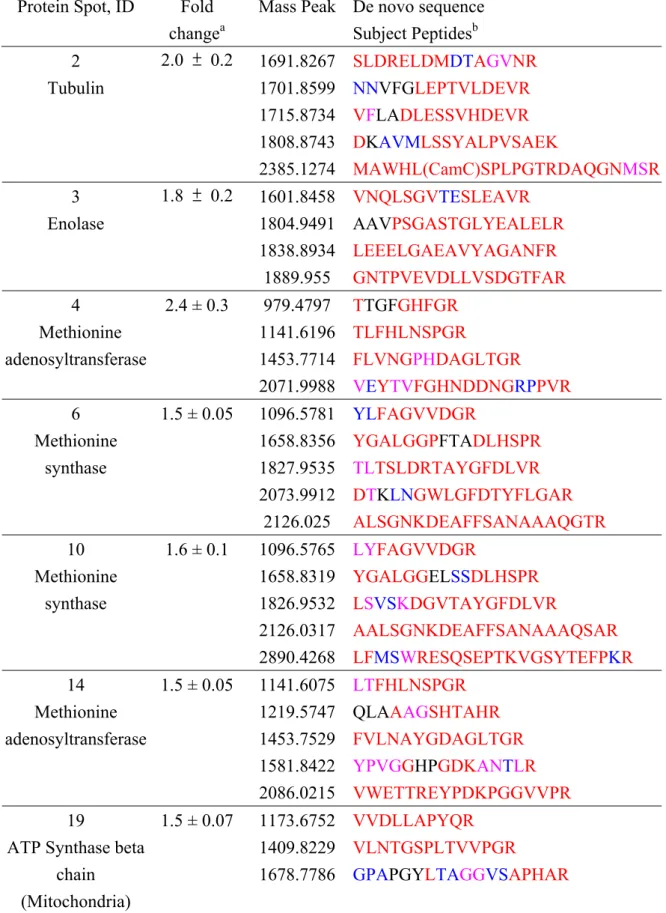
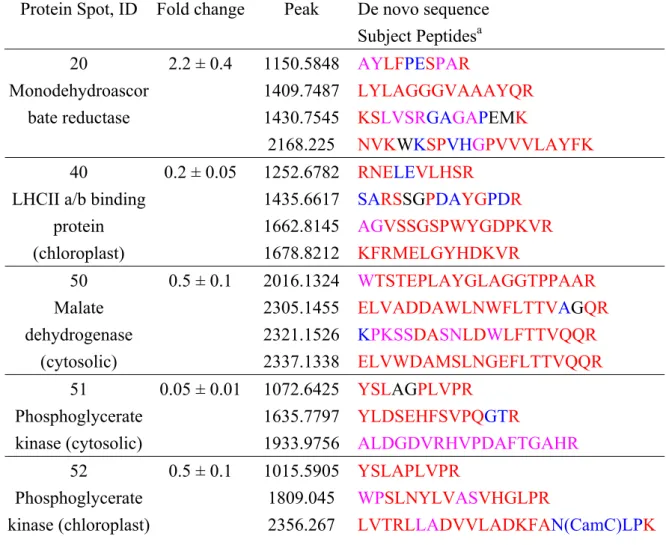
DryStrips was much better than pH 3-10; therefore, the pH 4-7 and 4.5-5.5 IPG DryStrips were used in the subsequent 2-DE analyses (Fig. 3A). There were 31 up-regulated and 24 down-regulated protein spots which were consistent in all three repeated gels. After MALDI-quadrupole (Q)-TOF MS/MS analysis, eight of up-regulated (spot No. 2, 3, 4, 6, 10, 14, 19 and 20) and four of down-regulated proteins (spot No. 40, 50, 51 and 52) were successfully identified with significant hits (P<0.05) in MASCOT searching program, while the remaining were unidentified due to low MOWSE scores. Additionally, these proteins were further identified using de
novo sequencing software, PEAKS, as listed in Table 1. The fold changes of each
protein spot are showed in Fig. 3B and Table 1. These proteins are involved in cell growth (spot No. 2, 4, 6, 10, 14), anti-stresses (spot No. 20), and cell metabolism including photosynthesis and respiration (spot No. 3, 19, 40, 50, 51, 52).
3.3 Functional classification of differentially expressed proteins
Using literature mining approach, we found these proteins involved in cell growth are tubulin (spot No. 2), S-adenosylmethionine synthase (SAM synthase, spot No. 4 and 14) and Methionine synthase (spot No. 6 and 10). There is one protein, Monodehydroascorbate reductase (MDAR, spot No. 20) involved in anti-stresses; six proteins relative to cell metabolism including enolase (spot No. 3), ATP synthase (spot No. 19), light-harvesting chlorophyll (LHCII) a/b binding protein (spot No. 40), Malate dehydrogenase (Malate DH, spot No. 50), cytosolic phosphoglycerate kinase (PGK, spot No. 51), and chloroplast phosphoglycerate kinase (spot No. 52).
3.3.1 Proteins involved in cell growth
Tubulin genes are often referred to as “housekeeping” genes. In plants, however, a more accurate label would be “house-building” gene, which reflects the importance of
various microtubule arrays in establishing patterns of cell elongation and cell division [26]. The importance of tubulin on BR-mediated cell growth has been proved in the reported study. DIM1 and BUL1 are two important genes involved in tubulin-based microtubules synthesis, after BR treatment of Arabidopsis dim1 and bul1 mutants, microtubules were reorganized and became correctly oriented [27]. In the assay of rice lamina inclination, tubulin was identified with evident increase under BR treatment [28]. Concluding theses findings, alpha-tubulin (spot No. 2) may be considered as a direct downstream component of BR’s action on epicotyl cell elongation in this study.
Among the proteinogenic amino acids, methionine (Met) displays many direct or indirect essential functions in cellular metabolism. Approximately 20 % of Met is incorporated into proteins and the remaining 80 % is converted to SAM, required for the biosynthesis of phenylpropanoid constituents of the cell wall, by SAM synthetase [29]. In the investigation of Arabidopsis seedlings growth, methionine synthase and SAM synthetase had a synchronized increase [30]. In this study, two methionine synthases (spot No. 6, 10) and two SAM synthetases (spot No. 4, 14) were up-regulated by EBL and could be corresponded to the elongated epicotyl.
In order to further elucidate the possible involvement of methionine synthesis on EBS-promoted epicotyl elongation, a specific methionine synthesis inhibitor, DL-propargylglycine (PAG) was used. Mung bean epicotyls, whose growth was initially suppressed by PAG partly, recovered their ability to elongate after treatment with EBL (Fig. 4). EBL-promoted epicotyl elongation may be regulated by inducing methionine biosynthetic pathway. The importance of methionine in cell growth has been reported for both plant and animal cells, particularly for the human cancer cells [31,32]; however, the research concerning the relationship between BR’s action and methionine’s is deficient. Our results may provide a possible correlation between
EBL-promoted epicotyl elongation and methionine biosynthetic pathway [33].
3.3.2 Protein involved in anti-stresses
Effects of BR anti-stresses, including temperature stress, salt stress, drought stress, and pathogen attack, have been preliminary investigated [6,33]. MDAR is an FAD enzyme which catalyzes the reduction of Monodehydroascorbate (MDA) radical to regenerate ascorbate using NAD(P)H as an electron donor. In plant cells, ascorbate is a major antioxidant that may act as a direct free radicals scavenger. The functional analysis of MDAR was studied in pea plants grown under eight stress conditions, including continuous light, high light intensity, continuous dark, mechanical wounding, low and high temperature, cadmium, and the herbicide 2,4-dichlorophenoxyacetic acid [34]. Thus, the up-regulation of MDAR (spot No. 20) in this study and our previous study [33] implies that the BR-mediated stress response in mung bean might also partially be regulated by MDAR.
3.3.3 Proteins involved in cell metabolism.
Two major metabolic pathways that energize plant cells are respiration and photosynthesis. In our study, we found out six proteins that involved in both pathways were regulated by BR, such as cytosolic PGK, chloroplast PGK, enolase, ATP synthase, LHCII a/b binding protein, and Malate DH. Cytosolic PGK, chloroplast PGK and enolase are responsible for triggering glycolysis; the ATP synthase is responsible for ATP production in the mitochondria; LHCII a/b binding protein is the main light harvesting antenna and important in photosynthesis of plants; and Malate DH is a kind of Malic enzymes (MEs). MEs catalyze the oxidative decarboxylation of L-malate, producing pyruvate, CO2, and NAD(P)H in the presence of a divalent cation [35]. This enzyme is widely distributed in nature due to the participation of the
reaction products in a large number of metabolic pathways [36].
To clarify if enolase is very important in BR-treated mung beans, we used enolase inhibitor, potassium fluoride (KF), to block enolase enzyme activity [37,38] and show that EBL can control the growth inhibition by KF. As shown in Fig. 4, the retarded elongation caused by KF reveals that enolase was important for cell growth; besides, the growth recovered after EBL treatment. These results showed a correlation between enolase and BR-promoted epicotyl elongation in mung bean. Briefly, the inhibitory epicotyl elongation by KF is caused by inhibiting enolase activity; however, EBL can induce reversely the epicotyl elongation of mung bean treated with enolase inhibitor, KF.
3.4 Discussion on potential molecular mechanism of EBL treatment: the
association between light intensity and EBL on epicotyl elongation
It has been reported that growth of the mung bean (Vigna radiata L.) epicotyl is retarded by white (400–700 nm) light, especially by monochromatic red (660 nm) light [1]. Growth promoting effects of BR are observed under those light conditions that retarded growth, but are not evident in the dark or under far-red light: BR’s effect seems to overcome the inhibitory affects of lights [39]. Nevertheless, the precise function of BR under the control of photomorphogenesis and light-regulated gene or protein expression is still unclear. In this study, a possible model of light affects BR’s action was proposed according to the proteomics results. As shown in Fig. 5, BR treatment decreased the synthesis of both cytosolic and chloroplastic PGK, which implied that under dark condition, glycolysis would cease due to the deficiency of 3-PG in cytosol. To facilitate the ongoing of glycolysis and ATP production, BR treatment must be held under light condition, so that the redundancy of chloroplastic 3-PG could transport into cytosol via an outer envelope membrane protein, OEP21
[40]. To prove this hypothesis, the 5-day-old mung bean seedlings treated with EBL were subjected to growth chambers with different light intensities. Consistent with our prediction, EBL-treated seedlings showed an elongated epicotyl under light condition (200 μmolm-2s-1), and the epicotyl elongation was reduced following the decreased light intensity (40 % light, 80 μmolm-2s-1). Moreover, EBL inhibited epicotyl elongation under full darkness condition (Fig. 6). In the latest study of BR signaling modulating phototropism of bak1 mutant in Arabidopsis, it also shows that the supplemental BL enhances high-light phototropism (100 μmolm-2s-1) and slightly reduces very-low-light phototropism (0.01 μmolm-2s-1) [41]. As a result, this study provides additional insight in understanding the integration of light and BR in elongating cells upon earlier studies [39]. Additionally, it also provides a possible molecular mechanism of EBL treatment.
4. Conclusions
Without doubts, the EBL acts as a growth-promoting compound to induce the elongation of mung beans epicotyls. In short, with de no sequencing, we successfully identified the differential proteins whose expression is altered by EBL. It may help us understand the biology of mung bean and the molecular mechanisms involved in response to EBL treatment. This study confirms that proteomic analysis is a powerful tool for evidencing the protein expression related to the effect of steroid hormones on epicotyl growth. Our approach opens a novel way of defining hormonal targets for growth of plants. This not only may illuminate a molecular mechanism of BR regulation but may also be further applied in plant breeding.
Acknowledgements
We specially acknowledge Hsuan-Cheng Huang (National Yang-Ming University, Taipei, Taiwan) for fruitful discussions and suggestions. The Core Facility for Proteomics (Academia Sinica, Taipei, Taiwan) is acknowledged. We also thank Shing-Chuan Chang for proteomics technical support, Ling- Long Huang for light microscopy support, Nancy Lin, Jason Lee and Derek Yang for manuscript proofreading. This work was supported by National Science Council of Taiwan (NSC 92-2311-B-002-029, NSC 93-2311-B-002-009 and NSC 94-3112-B-002-011).
References
[1] Clouse, S. D. & Sasse, J. M. (1998). BRASSINOSTEROIDS: Essential Regulators of Plant Growth and Development. Annu. Rev. Plant Physiol. Plant
Mol. Biol., 49, 427-451.
[2] Schumacher, K. & Chory, J. (2000). Brassinosteroid signal transduction: still casting the actors. Curr. Opin. Plant Biol., 3, 79-84.
[3] Bishop, G. J. & Koncz, C. (2002). Brassinosteroids and plant steroid hormone signaling. Plant Cell, 14 Suppl, S97-110.
[4] Wang, Z.Y. & He, J.X. (2004) Brassinosteroid signal transduction--choices of signals and receptors. Trends Plant Sci., 9, 91-96.
[5] Vert, G. & Chory, J. (2006). Downstream nuclear events in brassinosteroid signaling. Nature, 441, 96-100.
[6] Krishna, P. (2003). Brassinosteroid-Mediated Stress Responses. J. Plant
Growth Regul., 22, 289-297.
[7] Rao, S. S. R., Vardhini, B. V., Sujatha, E., & Anuradha, S. (2002) Brassinosteroids – A new class of phytohormones. Curr. Sci., 82, 1239-1245. [8] Grove, M. D., Spencer, G. F., Rohwedder, W. K., et al. (1979). Brassinolide, a
plant growth-promoting steroid isolated from Brassica napus pollen. Nature,
281, 216-217.
[9] Gregory, L. E. & Mandava, N. B. (1982). The activity and interaction of brassinolide and gibberellic acid in mung bean epicotyls. Physiol. Plant, 54, 239-243.
[10] Clouse, S. D., Zurek, D. M., McMorris, T. C., et al. (1992). Effect of Brassinolide on Gene Expression in Elongating Soybean Epicotyls. Plant
Physiol., 100, 1377-1383.
[11] Zurek, D. M. & Clouse, S. D. (1994). Molecular cloning and characterization of a brassinosteroid-regulated gene from elongating soybean (Glycine max L.) epicotyls. Plant Physiol., 104, 161-170.
[12] Goda, H., Shimada, Y., Asami, T., et al. (2002). Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol., 130, 1319-1334.
[13] Mussig, C., Fischer, S. & Altmann, T. (2002). Brassinosteroid-regulated gene expression. Plant Physiol., 129, 1241-1251.
[14] Wilkins, M. R., Sanchez, J. C., Gooley, A. A., et al. (1996). Progress with proteome projects: why all proteins expressed by a genome should be identified and how to do it. Biotechnol. Genet. Eng. Rev., 13, 19-50.
[15] Rampitsch, C., Bykova, N. V., McCallum, B., et al. (2006). Analysis of the wheat and Puccinia triticina (leaf rust) proteomes during a susceptible
host-pathogen interaction. Proteomics, 6, 1897-1907.
[16] Russeth, K. P., Higgins, L. & Andrews, M. T. (2006). Identification of proteins from non-model organisms using mass spectrometry: application to a hibernating mammal. J. Proteome Res., 5, 829-839.
[17] Habermann, B., Oegema, J., Sunyaev, S., et al. (2004). The power and the limitations of cross-species protein identification by mass spectrometry-driven sequence similarity searches. Mol. Cell. Proteomics, 3, 238-249.
[18] Jorge, I., Navarro, R. M. Lenz, C., et al. (2005) The holm oak leaf proteome: analytical and biological variability in the protein expression level assessed by 2-DE and protein identification tandem mass spectrometry de novo sequencing and sequence similarity searching. Proteomics, 5, 222-234.
[19] Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680-685.
[20] Juan, H. F., Chang, S. C., Huang, H. C., et al. (2005). A new application of microwave technology to proteomics. Proteomics, 5, 840-842.
[21] Onnerfjord, P., Ekstrom, S., Bergquist, J., et al. (1999). Homogeneous sample preparation for automated high throughput analysis with matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry. Rapid Commun. Mass
Spectrom., 13, 315-322.
[22] Ma, B., Zhang, K., Hendrie, C., et al. (2003). PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun.
Mass Spectrom., 17, 2337-2342.
[23] Nesvizhskii, A. I., Roos, F. F., Grossmann, J., et al. (2006). Dynamic spectrum quality assessment and iterative computational analysis of shotgun proteomic data: toward more efficient identification of post-translational modifications, sequence polymorphisms, and novel peptides. Mol. Cell. Proteomics, 5, 652-670.
[24] Frank, A., Tanner, S., Bafna, V., et al. (2005). Peptide sequence tags for fast database search in mass-spectrometry. J. Proteome Res., 4, 1287-1295.
[25] Symons, G. M. & Reid, J. B. (2004). Brassinosteroids do not undergo long-distance transport in pea. Implications for the regulation of endogenous brassinosteroid levels. Plant Physiol., 135, 2196-2206.
[26] Munoz, F. J., Labrador, E. & Dopico, B. (1998). Brassinolides promote the expression of a new Cicer arietinum beta-tubulin gene involved in the epicotyl elongation. Plant Mol. Biol., 37, 807-817.
[27] Catterou, M., Dubois, F., Schaller, H., et al. (2001). Brassinosteroids, microtubules and cell elongation in Arabidopsis thaliana. II. Effects of brassinosteroids on microtubules and cell elongation in the bul1 mutant.
Planta, 212, 673-683.
[28] Konishi, H. & Komatsu, S. (2003). A proteomics approach to investigating promotive effects of brassinolide on lamina inclination and root growth in rice seedlings. Biol. Pharm. Bull., 26, 401-408.
[29] Hesse, H., Kreft, O., Maimann, S., et al. (2004). Current understanding of the regulation of methionine biosynthesis in plants. J. Exp. Bot., 55, 1799-1808. [30] Gallardo, K., Job, C., Groot, S.P., et al. (2002). Importance of methionine
biosynthesis for Arabidopsis seed germination and seedling growth. Physiol.
Plant, 116, 238-247.
[31] Hesse, H. & Hoefgen, R. (2003). Molecular aspects of methionine biosynthesis. Trends Plant Sci., 8, 259-262.
[32] Cellarier, E., Durando, X., Vasson, M. P., et al. (2003). Methionine dependency and cancer treatment. Cancer Treat. Rev., 29, 489-499.
[33] Huang, B., Chu, C. H., Chen, S. L., et al. (2006). A proteomics study of the mung bean epicotyl regulated by brassinosteroids under conditions of chilling stress. Cell. Mol. Biol. Lett., 11, 264-278.
[34] Leterrier, M., Corpas, F. J., Barroso, J. B., et al. (2005). Peroxisomal monodehydroascorbate reductase. Genomic clone characterization and functional analysis under environmental stress conditions. Plant Physiol., 138, 2111-2123.
[35] Chang, G. G. & Tong, L. (2003). Structure and function of malic enzymes, a new class of oxidative decarboxylases. Biochemistry, 42, 12721-12733.
[36] Lance, C. & Rustin, P. (1984). The central role of malate in plant metabolism.
Physiol. Veg., 22, 625–641.
[37] Saier, M. H., Jr. & Feucht, B. U. (1980). Regulation of carbohydrate transport activities in Salmonella typhimurium: use of the phosphoglycerate transport system to energize solute uptake. J. Bacteriol., 141, 611-617.
[38] Otani, M., Ihara, N., Umezawa, C., et al. (1986). Predominance of gluconate formation from glucose during germination of Bacillus megaterium QM B1551 spores. J. Bacteriol., 167, 148-152.
[39] Clouse, S.D. (2001). Integration of light and brassinosteroid signals in etiolated seedling growth. Trends Plant Sci., 6, 443-445.
[40] Bolter, B., Soll, J., Hill, K., et al. (1999). A rectifying ATP-regulated solute channel in the chloroplastic outer envelope from pea. EMBO J., 18, 5505-5516.
[41] Whippo, C.W. & Hangarter, R.P. (2005). A brassinosteroid-hypersensitive mutant of BAK1 indicates that a convergence of photomorphogenic and hormonal signaling modulates phototropism. Plant Physiol., 139, 448-457.
Figure legends
Figure 1. Effect of 24-epibrassinolide (EBL) on epicotyl elongation. The 5-day-old
mung bean seedlings treated with 10μM EBL onto shoot meristem were subjected to growth chamber for one day (A), and the different elongation responses of hypocotyl and epicotyl to EBL treatment were compared (D). In order to monitor the elongation effect by EBL, mung bean epicotyl was divided into three sections as indicated (B and E). Ten micromoles EBL was also applied to 10-day-old soybean seedlings (C) and the epicotyl elongation rate between mung bean and soybean were compared (F). Data are shown as the mean ± s.d. Statistic bars marked with the same letter are not significantly different by Fishers LSD test (p<0.05). Bar = 2 cm.
Figure 2. Epicotyl cell expansion was promoted by EBL. The epicotyl sections (2mm
length from meristem) from EBL or control treatments were excised for light microscopy observation. Semi-thin sections with 1μm thickness were prepared by ultramicrotome and stained with toluidine blue. A: cross-section, B: longisection, P: pith, V: vascular, C: cortex, E: epidermis. Bar = 100 μm.
Figure 3. 2-DE analysis of epicotyl proteins regulated by EBL. Three hundred
micrograms of epicotyl proteins with EBL or mock treatments were separated by different pH-based (pH 4.5-5.5 and pH 4-7) 2-DE analyses (A). Arrows in the control treatment represented proteins that were down-regulated by EBL, and the arrows in the EBL treatment represented proteins that were up-regulated by EBL. Twelve annotated proteins regulated by EBL in Table 1 were focused respectively (B). Relative fold changes were calculated statistically from three repeats.
elongation. The 5-day-old mung bean seedlings were treated with EBL, or not (control), and/or various inhibitors, for two days and determined the phenotypic differences among treatments respectively. The abbreviation and concentration of inhibitors were illustrated as: DL-propargylglycine, PAG, 10μM; potassium fluoride, KF, 10μM. PAG/EBL, and KF/EBL represented that after inhibitors treatment for 30 min., seedlings were treated with EBL. Data are shown as the mean ± s.d. Statistic bars marked with the same letter are not significantly different by Fishers LSD test (p<0.05).
Figure 5. A possible mechanism of light effect on BR-promoted epicotyl elongation.
A possible model of light affects BR’s action was proposed according to the proteomics results. Exogenous treatment of EBL decreases the synthesis of both chloroplast and cytosolic phosphoglycerate kinase (PGK). High light exposure to the seedlings contributes to the accumulation of the 3-PG in chloroplast, which then causes redundant chloroplast 3-PG to be transported into cytosol thorough a hypothetic outer membrane protein, OEP 21, and would subsequently be catalyzed by an up-regulated enolase. Enolase stimulates downstream pyruvate synthesis and thus continues the glycolytic pathway. Lastly, bulk ATP would be produced by up-regulated ATP synthase in the mitochondria, and the epicotyl elongation is observed.
Figure 6. Effect of light intensity on EBL-mediated epicotyl elongation. To prove the
hypothesis in Fig. 5, the 5-day-old mung bean seedlings treated with EBL were subjected to growth chambers with different light intensities. The 5-day-old mung bean seedlings treated with 10μM EBL or equal amount of solvent (control) were subjected to different light intensities for one day (A). Comparison of epicotyls length
in (A) is shown in (B). Data are shown as the mean ± s.d. Statistic bars marked with the same letter are not significantly different by Fishers LSD test (p<0.05) (C). Bar = 2 cm.
Table 1. Homology-Based Search Results. PEAKS de novo Sequencing and MS BLAST
Database Search Results Protein Spot, ID Fold
changea
Mass Peak De novo sequence Subject Peptidesb 1691.8267 SLDRELDMDTAGVNR 1701.8599 NNVFGLEPTVLDEVR 1715.8734 VFLADLESSVHDEVR 1808.8743 DKAVMLSSYALPVSAEK 2 Tubulin 2.0 ± 0.2 2385.1274 MAWHL(CamC)SPLPGTRDAQGNMSR 1601.8458 VNQLSGVTESLEAVR 1804.9491 AAVPSGASTGLYEALELR 1838.8934 LEEELGAEAVYAGANFR 3 Enolase 1.8 ± 0.2 1889.955 GNTPVEVDLLVSDGTFAR 979.4797 TTGFGHFGR 1141.6196 TLFHLNSPGR 1453.7714 FLVNGPHDAGLTGR 4 Methionine adenosyltransferase 2.4 ± 0.3 2071.9988 VEYTVFGHNDDNGRPPVR 1096.5781 YLFAGVVDGR 1658.8356 YGALGGPFTADLHSPR 1827.9535 TLTSLDRTAYGFDLVR 2073.9912 DTKLNGWLGFDTYFLGAR 6 Methionine synthase 1.5 ± 0.05 2126.025 ALSGNKDEAFFSANAAAQGTR 1096.5765 LYFAGVVDGR 1658.8319 YGALGGELSSDLHSPR 1826.9532 LSVSKDGVTAYGFDLVR 2126.0317 AALSGNKDEAFFSANAAAQSAR 10 Methionine synthase 1.6 ± 0.1 2890.4268 LFMSWRESQSEPTKVGSYTEFPKR 1141.6075 LTFHLNSPGR 1219.5747 QLAAAGSHTAHR 1453.7529 FVLNAYGDAGLTGR 1581.8422 YPVGGHPGDKANTLR 14 Methionine adenosyltransferase 1.5 ± 0.05 2086.0215 VWETTREYPDKPGGVVPR 1173.6752 VVDLLAPYQR 1409.8229 VLNTGSPLTVVPGR 19 ATP Synthase beta
chain (Mitochondria)
1.5 ± 0.07
Table 1. Continued
Protein Spot, ID Fold change Peak De novo sequence Subject Peptidesa 1150.5848 AYLFPESPAR 1409.7487 LYLAGGGVAAAYQR 1430.7545 KSLVSRGAGAPEMK 20 Monodehydroascor bate reductase 2.2 ± 0.4 2168.225 NVKWKSPVHGPVVVLAYFK 1252.6782 RNELEVLHSR 1435.6617 SARSSGPDAYGPDR 1662.8145 AGVSSGSPWYGDPKVR 40 LHCII a/b binding
protein (chloroplast) 0.2 ± 0.05 1678.8212 KFRMELGYHDKVR 2016.1324 WTSTEPLAYGLAGGTPPAAR 2305.1455 ELVADDAWLNWFLTTVAGQR 2321.1526 KPKSSDASNLDWLFTTVQQR 50 Malate dehydrogenase (cytosolic) 0.5 ± 0.1 2337.1338 ELVWDAMSLNGEFLTTVQQR 1072.6425 YSLAGPLVPR 1635.7797 YLDSEHFSVPQGTR 51 Phosphoglycerate kinase (cytosolic) 0.05 ± 0.01 1933.9756 ALDGDVRHVPDAFTGAHR 1015.5905 YSLAPLVPR 1809.045 WPSLNYLVASVHGLPR 52 Phosphoglycerate kinase (chloroplast) 0.5 ± 0.1 2356.267 LVTRLLADVVLADKFAN(CamC)LPK
a Fold change was determined as EBL/Control and the mean ± s.d from three repeats bThe confidence level for each individual amino acid in the sequence using different
出席國際學術會議心得報告
計畫編號 NSC95-2311-B-002-007 計畫名稱 油菜固醇對綠豆上胚軸延長及低溫反應的蛋白質體與代謝質體研究 出國人員姓名 服務機關及職稱 陳益明 / 台灣大學植物科學研究所 / 名譽教授 會議時間地點 2007 年 7 月 7 日至 11 日 / 美國芝加哥會議名稱 全美國植物生物學會(American Society of Plant Biologists) 2007 年會暨學術 研討會
發表論文題目 Transcriptome and Proteome Change of Brassinosteroid Biosynthetic Gene and Down-stream Proteins under Chilling Condition in Mung Bean Seedlings
一、參加會議經過
全美國植物生物學會(American Society of Plant Biologists) 2007 年聯合其他三個美國植物科學 會,包括 American Fern Society、American Society of Plant Taxonomists 及 Botanical Society of America 於 7 月 7 日至 11 日在美國芝加哥 Hilton 大飯店舉行聯合年會暨學術研討會,與會之 學者、專家及產業界代表共有數千人,盛況空前。 全美國植物生物學會之前身為全美國植物生理學會,成立於 1924 年,它是全球最早成立及會 員最多的植物科學相關學會。本人於 1973 年就加入此學會,至今已有 35 年之久。由於植物 科學近二十年來發展神速,各國政府及科學家都相爭投入現代植物科學的基礎與應用之研 究,更有許多產業界投入於遺傳工程等研究。因此於 2001 年才更名為全美國植物生物學會。 該學會每年在美國各州及加拿大舉辦年會及學術研討會,1973 年 7 月本人首次參加該學會之 年會,當年是在康乃爾大學舉行,當時參加的學者有林秋榮院士、本人及現任中研院植微所 所長賀端華院士,當年他還是密西根州立大學博士班之研究生。該學會每年出版全球最傑出 的植物科學相關學術期刊及 Review 型式的出版品,例如 Plant Cell (Impact factor: 9.865)、Plant Physiology (IP: 6.125)及 Annual Review of Plant Biology (IP: 19.837)等。
今年的主題包括 Plant Biology in Sub-Saharan Africa, Plant and Human Nutrition, Emerging Genome Technologies, Vegetative Development, Cell-to-cell and Long Distance Signaling, Celluler Technologists, Abiotic Stress, Rhythms, Seed Biology, Education and Outreach, Plant Defense, Protein Targeting,
Heavy Metals and Phytomediation, Pollen Biology, Cell Walls, Envivonmental Signaling, Evolution of Development and Physiology, Plant Symbiont Biology, Mechanisms of Gene Regulation, Evolutionary Development, Manipulation of Host Signaling by Pathogens 等 21 個主題,本人在此年會中以壁報方 式參加展示,其題目是 ”Transcriptome and Proteome Change of Brassinosteroid Biosynthetic Gene and Down-stream Proteins under Chilling Condition in Mung Bean Seedlings”。在本報告中展示綠豆
CYP90A2 是油菜類固醇合成的一個重要基因,它與阿拉伯芥的 CPD 基因有 77%的相同性,該 基因有下列四項特性:(1)在幼苗發育過程,葉部的表現會增加;(2)顯現 Diurnal 變化;(3)BR 會 feedback regulated;(4)在 10℃低溫下會顯著抑制其表現。若外加 BR,受低溫抑制的生長及 存活率都會受到顯著改善。在低溫逆境下,BR 促進上胚軸的延長,藉蛋白質體技術的分析研 究,其中有 31 的蛋白質點是增加的,但亦有 24 個蛋白質點是減少表現的。其中有 17 個受低 溫逆境而減少表現的蛋白質亦會因外加 BR 而轉變為增加。以 MALDI-Q-TOF MS/MS 質譜分析 鑑定這些蛋白質,由結果顯示這些蛋白質大都參與細胞生長,光合作用及呼吸作用等。藉生 理檢定有兩項新發現:(1)只在強光下 BR 才能促進上胚軸的延長;(2)低溫逆境能抑制綠豆上 胚軸的延長,但外加 DL-methionine 能部分恢復。由上述研究可了解 brassinosteroid 與低溫適應 二者間的相互作用機裡。 二、與會心得 本次參加了許多場的研討會,包括中研院分生所李秀敏博士主講的”Stimulation of trasitpeptide Release and ATP hydrolysis by a Cochaperone”,她英文極為流利,內容新穎,是國內表現傑出 的年輕科學家,應值得鼓勵!
本次研討會因與其他三個學會一起舉辦,因此會場安排不佳,服務人員不足,演講廳又不容 易找,造成會場較混亂是美中不足之處。本次參加會議,攜帶回來的資料有 2007 年 Plant Biology and Botany 的 Program & Abstract Book 及 Poster & Abstract Book 等資料。
感謝行政院國家科學委員會常年來的經費補助,我雖已退休教職,但使我這 35 年來能持續不 斷地參加該學會的年會及學術研討會,其意義重大。