Molecular epidemiology of invasive Candida albicans at a tertiary hospital in northern Taiwan from 2003 to 2011
Shao-Hung Wang1, Mandy Shen1, Hsin-Chieh Lin2, Pei-Lun Sun3, Hsiu-Jung Lo4,5,
Jang-Jih Lu2,6*
1Department of Microbiology, Immunology and Biopharmaceuticals, National Chiayi
University, Chiayi City, Taiwan
2Department of Laboratory Medicine, Chang Gung Memorial Hospital, Linkou,
Taoyuan, Taiwan
3Department of Dermatology, Chang Gung Memorial Hospital, Linkou, Taoyuan,
Taiwan
4National Institute of Infectious Diseases and Vaccinology, National Health Research
Institutes, Miaoli, Taiwan
5School of Dentistry, China Medical University, Taichung, Taiwan
6Department of Medical Biotechnology and Laboratory Science, Chang Gung
University, Taoyuan, Taiwan Running title:
Molecular epidemiology of invasive C. albicans in Taiwan
*Address correspondence to: Dr. Jang-Jih Lu, MD, PhD.
Department of Laboratory Medicine, Chang Gung Memorial Hospital, 5 Fu-Shing St., Kwei-Shan, Taoyuan 333, Taiwan.
Phone: 886-3-328-1200 ext. 2554 Fax: 886-3-397-1827 E-mail: janglu45@gmail.com 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25
Abstract
Candida albicans is a common cause of bloodstream fungal infections in
hospitalized patients. To investigate its epidemiology, multilocus sequence typing (MLST) was performed on 285 C. albicans bloodstream isolates from patients in Chang Gung Memorial Hospital at Linkou (CGMHL), Taiwan from 2003 to 2011. Among these isolates, the three major diploid sequence types (DSTs) were 693, 659, and 443 with 19, 16, and 13 isolates, respectively. The 179 DSTs were classified into 16 clades by unweighted pair-group method using arithmetic averages (UPGMA). The major ones were clades 1, 4, 3 and 17 (54, 49, 31 and 31 isolates, respectively). Further analyses with eBURST clustered the 285 isolates into 28 clonal complexes (CC). The most common complexes were CC8, CC20, and CC9. DST 693 that had the highest number of isolates was determined to be the cluster founder of CC20, which belonged to clade 3. So far, 33 isolates worldwide including 29 from Taiwan and 4 from Korea, are CC20, suggesting that CC20 is an Asian cluster. Two
fluconazole-resistant isolates belonging to CC12 and CC19 were detected. All other CGMHL isolates were susceptible to 5-flucytosine, amphotericin B, anidulfungin, caspofungin, fluconazole, itraconazole, micafungin, posaconazole, and voriconazole. However, CC20 isolates exhibited significantly lower susceptibility to fluconazole. In conclusion, the 285 CGMHL C. albicans isolates displayed geographically clustering with Asian isolates, and most of them are susceptible to common antifungal drugs. Isolates of DST 693, a Taiwanese major genotype belonging to MLST clade 3, were more resistant to fluconazole than other isolates.
Keywords: Candida albicans, candidemia, multilocus sequence typing (MLST), antifungal susceptibility test
26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50
Introduction
Candida albicans is an organism of the normal gut flora in humans. It is also an
opportunistic pathogen and is the fourth most common cause of nosocomial bloodstream infections with a mortality rate of 30-60%.1-5 The risk factors of candidemia include invasive surgeries such as dialysis,6 implantation of central venous catheter,7 diabetes,6 burns,8 HIV infections, immunosuppression due to chemotherapy,9 and use of steroid drugs or broad-spectrum antibiotics.7
The increasing frequency of invasive candidiasis and its serious outcome demand more epidemiological studies. A well-accepted method for typing C. albicans isolates is the multilocus sequence typing (MLST).10-12 It is based on nucleotide sequence variations within the 300- to 400-bp internal regions of seven housekeeping genes, including AAT1a, ACC1, ADP1, MPIb, SYA1, VPS13, and ZWF1b. A sequence variation in each locus is assigned an allele number. Combination of allele numbers of these seven genes constitutes a unique diploid sequence type (DST) of a C. albicans isolate. Because MLST analysis relies only on nucleotide sequencing, the information about C. albicans isolates can be exchanged around the world through a global
database (http://calbicans.mlst.net).12,13 At least 2400 DSTs have been recorded in the C. albicans MLST database.
UPGMA (unweighted pair-group method using arithmetic averages) is a method that can be used to determine the phylogenetic relationship among C. albicans
isolates.11,14 Using this method, Gong et al. has recently classified 1500 C. albicans
isolates into 18 distinct clades.14 Clade 1 isolates distribute globally, whereas isolates from the Pacific Rim cluster mostly in clades 14 and 17. Clade 1 isolates in general have a higher acid phosphatase activity and are less susceptible to 5-fluorocytosine11,15
and more salt tolerant16. Another method called eBURST (electronic Based Upon 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75
Related Sequence Types) allows determination of patterns of evolutionary descent by grouping isolates that differ at one or two of the seven MLST alleles into clonal clusters.17
In this study, we determined the DSTs of 285 C. albicans isolates causing bloodstream infections in Chang Gung Memorial Hospital at Linkou (CGMHL) and investigated the epidemiology of the isolates using both UPGMA and eBURST. We also determined antifungal susceptibility of these isolates. Results showed that isolates of DST 693/clonal complex 20/clade 3 are more resistant to fluconazole than other isolates.
Materials and methods Candida albicans isolates
A total of 285 bloodstream infection isolates from CGMHL obtained between 2003-2011, including all 72 from pediatric patients and 213 randomly selected from 1098 archived adult ICU isolates, were investigated in this study (Supplementary Table). Each isolate was collected only once from a patient within the hospital admission. All isolates were identified by MALDI-TOF mass spectrometry and germ tube formation methods or CHROMagar Candida (BD).
Multilocus sequence typing
MLST of C. albicans isolates were performed as described by Bougnoux et al..12,13 A
portion of each of AAT1a, ACC1, ADP1, MPIb, SYA1, VPS13, and ZWF1b genes was amplified by PCR, and the resulting PCR products were sequenced. Each nucleotide sequence thus generated was compared to those in the C. albicans MLST database (http://calbicans.mlst.net/) to obtain an allele number. Any sequence that does not 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100
match with any of the preexisting sequences was given a new allele number. The combination of the 7 allele numbers defined a unique DST of an isolate.
UPGMA analysis
To determine phylogenetic relatedness, DSTs of the 285 CGMHL isolates and 996 isolates with known clades retrieved from the C. albicans MLST database were analyzed by UPGMA as described previously.11,14 Briefly, the sequences of each
housekeeping gene of each isolate were aligned to reveal polymorphic bases. The seven MLST polymorphic sequences from each isolate were then concatenated into a single sequence. Each base of the combined sequence was rewritten with two letters representing a homozygous or heterozygous diploid sequence. The genetic relatedness of the transformed sequences were analyzed by the software MEGA version 6 to generate a dendrogram.18
eBURST analysis
The relationships among the 285 CGMHL isolates and all 2448 isolates in the MLST database (date accessed 10.01.14) were determined by eBURST
(http://eburst.mlst.net/). Based on the 7 allele numbers of each isolate, eBURST placed related isolates into a clonal complex (CC) and predicted the ancestral DST of each CC by calculating the frequency of each DST genotype. The results of eBURST were displayed as the most parsimonious pattern of each descent of the ancestral DST type.
Antifungal susceptibility testing
A commercially available dried colorimetric microdilution panel (Sensititre 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125
YeastOne, TREK Diagnostic Systems) was used for susceptibility testing of C.
albicans isolates to 5-flucytosine, amphotericin B, anidulafungin, caspofungin,
fluconazole, itraconazole, micafungin, posaconazole, and voriconazole. Briefly, C.
albicans isolates (1.5 - 8 x 102 cfu) were seeded in YeastOne medium containing
antifungal agents and incubated at 35ºC without CO
2 for 24 hours. The minimum
inhibition concentration (MIC) of each antifungal agent was determined according to the guideline provided by the kit. The clinical breakpoints for sensitive, intermediate, and resistant isolates and epidemiological cutoff values for wildtype and non-wildtype isolates for the antifungal agents were referenced to those of Pfaller et al..19 MIC
50 and
MIC90 values of C. albicans isolates against each antifungal agent were also
calculated. Isolates with a fluconazole MICs >=0.5 µg/ml were defined as having a lower fluconazole susceptibility, i.e. more resistant to fluconazole.
Statistical analysis
The chi-square and Fisher exact tests were performed to compare genotype distributions and antibiotic susceptibility. A p-distance < 0.05 was considered significant. Prism 5.0 software (GraphPad, San Diego) was used for the analysis.
Results
Multilocus sequencing typing
Of the 285 isolates, 172 isolates (60.4%) were assigned 68 previously defined DSTs, and the other 113 isolates were assigned 110 new DSTs (DST 2427-2555) (Supplementary Table). The 213 isolates from adult patients were assigned 142 DSTs, and the 72 isolates from pediatric patients were assigned 53 different DSTs. Among the 213 isolates from adults, the most prevalent clones were DST 659 (combination 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150
alleles 11, 26, 6, 4, 34, 60, and 119 for AAT1a, ACC1, ADP1, MPIb, SYA1, VPS13, and ZWF1b, respectively), DST 693 (combination alleles 1, 7, 15, 6, 61, 105, and 112), DST 443 (combination alleles 59, 5, 21, 2, 80, 108, and 15), and DST 766 (combination alleles 23, 3, 5, 3, 57, 100, and 6). Among the 72 isolates from children, the most dominant clones were DST693, DST 1849 (combination alleles 5, 5, 5, 9, 2, 6, and 5), DST 365 (combination alleles 55, 14, 4, 3, 6, 45, and 15), DST 443, and DST 659. Overall, the three most prevalent DST clones were DST 693 (19 isolates; 6.7%), DST 659 (16 isolates; 5.6%), and DST 443 (13 isolates; 4.6%) (Supplementary Table).
MLST clade distribution of CGMHL C. albicans isolates
To compare the genotypes of the CGMHL isolates to those of global isolates with previously reported MLST clades, UPGMA phylogenetic analyses were
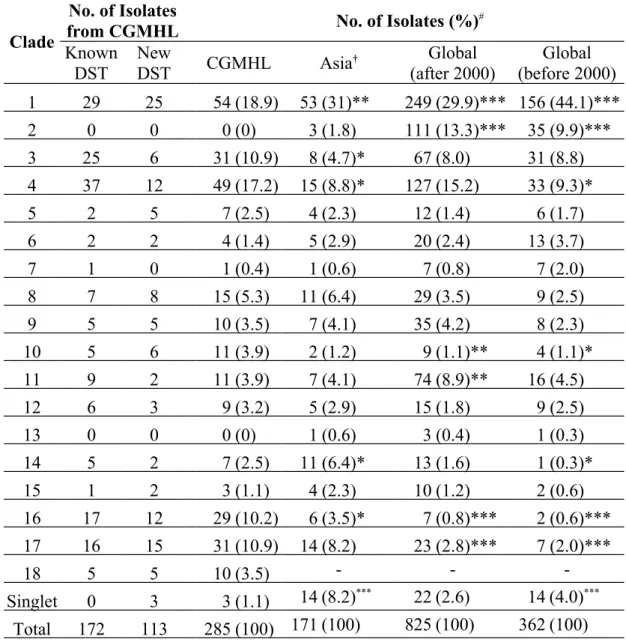
performed. The MLST clade distribution of all C. albicans isolates is shown in Figure 1 and Table 1, and the detail of MLST genotyping is shown in the Supplementary Table. The most common clades were clade 1 (54 isolates, 18.9%), clade 4 (49 isolates, 17.2%), clade 3 (31 isolates, 10.9%), clade 17 (31 isolates, 10.9%), and clade 16 (29 isolates, 10.2%). Among the 113 isolates with newly assigned DSTs, the most common clades were clade 1 (25 isolates, 22.1%), clade 17 (15 isolates, 13.3%), clades 4 and 16 (12 isolates each, 10.6%) (Table 1). Within the same clade, the isolates from this study were clustered together, especially those in clades 3, 4, and 11 (Figure 1).
eBURST clonal clustering of CGMHL C. albicans isolates
To determine whether the CGMHL isolates that clustered together within the 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175
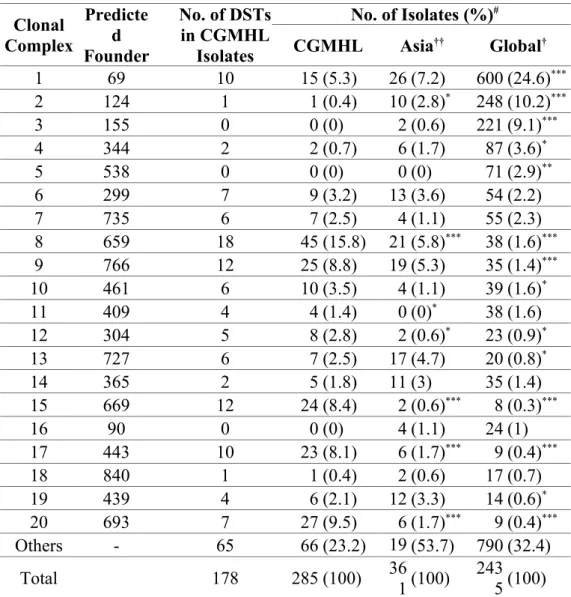
same clade belonged to a specific genotype, eBURST analysis was performed. The 285 CGMHL isolates were clustered in 28 clonal complexes (CCs) (Supplementary Table), and 195 (55.8%) isolates belonged to 6 CCs, including CC1, CC8, CC9, CC15, CC17 and CC20 (Table 2). The most common cluster was CC8 (45 isolates, 15.8%), followed by CC20 (27 isolates, 9.5%), and CC9 (25 isolates, 8.8%). The predicted clonal founders of CC8 and CC20 were DST 659 and DST 693 (Table 2), respectively. They were also the most dominant DSTs in this study. The majority of isolates of clades 3, 4, 11, 16, and 17 were clustered in CC20 (27/31, 87.1%), CC8 (45/49, 91.8%), CC10 (10/11, 90.9%), CC15 (24/29, 82.8%), and CC17 (23/31, 74.2%), respectively (Table 3).
Antifungal susceptibility testing of CGMHL C. albicans isolates
The mean MIC50 of the 285 CGMHL isolates against 5-flucytosine, amphotericin
B, anidulafungin, caspofungin, fluconazole, itraconazole, micafungin, posaconazole, and voriconazole were 0.06 µg/ml, 0.5 µg/ml, 0.06 µg/ml, 0.06 µg/ml, 0.5 µg/ml, 0.03 µg/ml, 0.008 µg/ml, 0.015 µg/ml, and 0.008 µg/ml, respectively. The mean MIC90 of
the 285 CGMHL isolates against these antifungal agents were 0.12 µg/ml, 0.5 µg/ml, 0.06 µg/ml, 0.06 µg/ml, 0.5 µg/ml, 0.06 µg/ml, 0.008 µg/ml, 0.03 µg/ml, and 0.008 µg/ml, respectively. Two fluconazole resistant isolates C001 (DST 1933/CC19/clade 14) and D034 (DST 1363/CC12/clade 10) from adult patients were detected
(Supplementary Table). The remaining isolates were susceptible to all 9 drugs tested. Most MIC50 and MIC90 values of the antifungals tested in CGMHL isolates are similar
to that reported in a global survey conducted in 2013.20 The MIC
50 and MIC90 of
micafungin in CGMHL isolates (0.008 µg/ml and 0.008 µg/ml, respectively) is lower than that in the survey (0.015 µg/ml and 0.03 µg/ml, respectively), but that of
176 177 178 179 180 181 182 183 184 185 186 187 188 189 190 191 192 193 194 195 196 197 198 199 200
fluconazole (MIC50=0.5 µg/ml and MIC90=0.5 µg/ml) and caspofungin (MIC50=0.06
µg/ml and MIC90=0.06 µg/ml) in this study are higher than the survey (fluconazole
MIC50=0.12 µg/ml and MIC90=0.25 µg/ml; caspofungin MIC50=0.03 µg/ml and
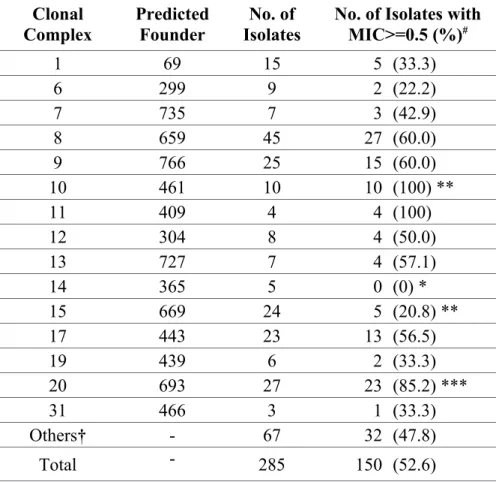
MIC90=0.03 µg/ml). One hundred and fifty isolates (52.6%) were found to be less
susceptible to fluconazole (MIC>=0.5 µg/ml), including all CC10 isolates and 85.2% of CC20 isolates (Table 4 and Supplementary Table).
Discussion
In the current study, the epidemiology of 285 bloodstream isolates of C.
albicans was investigated. Based on nucleotide sequence variations in the 7
housekeeping genes, 172 isolates were assigned 68 previously known DSTs; the other 113 (39.6%) isolates were assigned 110 new DSTs. The number of variable bases of each of the 7 housekeeping genes was 20 for VPS13; 18 for MPIb; 16 each for ADP1,
SYA1 and ZWF1b; 10 for AAT1a, and 6 for ACC1. The number of genotypes (alleles)
of each gene was 57 for VPS13, 39 for ZWF1b, 38 for SYA1, 34 for AAT1a, 28 for
MPIb, 21 for ACC1, and 19 for ADP1. Of the 7 genes used for MLST, ACC1 and AAT1a showed the highest typing efficiency, distinguishing 3.50 (21 genotypes
divided by 6 variable bases) and 3.40 (34 genotypes divided by 10 variable bases) genotypes per polymorphism, respectively. These two genes were also found to have the best discriminating power by Bougnoux et al..21
The 285 CGMHL and 996 MLST reference isolates were classified into 18 clades by UPGMA. The CGMHL isolates (open circles in Figure 1) were clustered in all MLST clades except clades 2 and 13. Isolates in these two clades are mostly found in Europe and Africa, and clade 13 was previously recognized as Candida africana by phenotying.11,22 Clades 1-4 and 11 have been evidenced the most consistent during
201 202 203 204 205 206 207 208 209 210 211 212 213 214 215 216 217 218 219 220 221 222 223 224 225
rapid expansion of the database, and clades 3 and 11 have been shown to be very close to each other.22 The uneven distribution of the CGMHL isolates in clades 3, 4, and 11 (Figure 1) suggests a close phylogenetic association of CGMHL isolates within the same clade. The population of CGMHL isolates in clade 1 and singlets (isolates that could not be classified into any clade by UPGMA with a cutoff value of
p=0.04) was much smaller but that in clade 16 was significantly bigger than that of
the global isolates published in 2007.11 The populations of CGMHL isolates in clades 3 and 4 were bigger than those of other Asian isolates, but not those of global isolates collected since 2000 (Table 1). In contrast, the number of CGMHL isolates in clade 17 was higher than that of global (both before and after 2000) but not of other Asian isolates.
UPGMA measures the p-distance of polymorphic nucleotide sequences.
Although it provides a simple view of phylogenetic relationship of the isolates, some minor clades were altered when the isolate number increased.22 Therefore, eBURST, another powerful algorism to reveal the genetic relationship of isolates, was also used in this study. Results showed that the percentages of CC8, CC20, CC15 and CC17 CGMHL isolates (15.8%, 9.5%, 8.4% and 8.1%, respectively) were significantly higher than those of other Asian (5.8%, 1.7%, 0.6% and 1.7%) and global isolates (1.6%, 0.4%, 0.3% and 0.4%), suggesting an expansion of CC8, CC20, CC15 and CC17 isolates in CGMHL (Table 2).
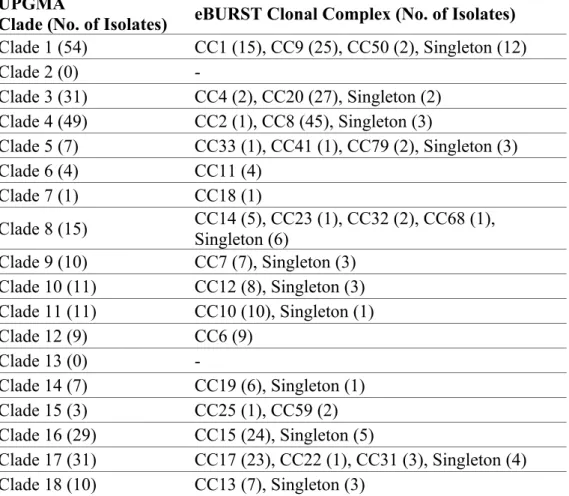
Results of this study also showed that the CGMHL isolates in the same eBURST clonal complexes were grouped together in the same UPGMA clades (Table 3). Thus, there was a good correlation between UPGMA grouping and eBURST clustering. DST 659 was the predicted founder of the largest CC in the CGMHL isolates. DST 659 was determined to be CC11 (i.e. 11th largest eBURST cluster) by Odds et al.,11 226 227 228 229 230 231 232 233 234 235 236 237 238 239 240 241 242 243 244 245 246 247 248 249 250
but was determined to be CC8 (i.e. 8th largest eBURST cluster) in this study
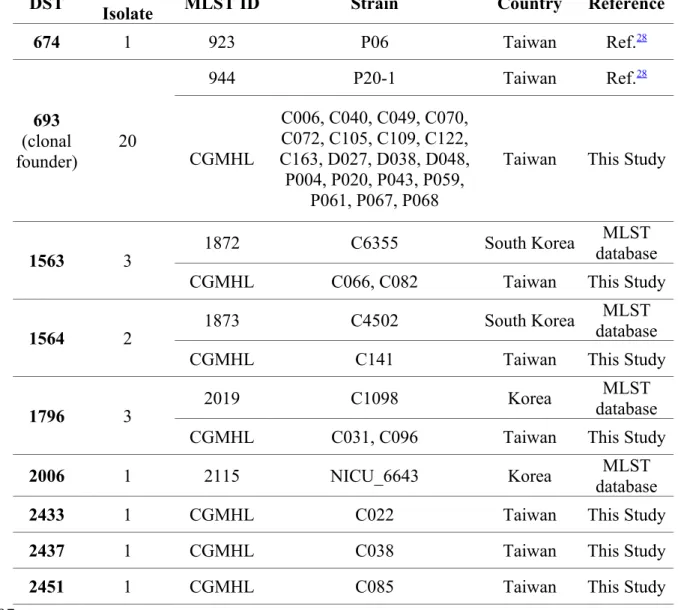
(Supplementary Table). In addition, DST 693, which was previously classified as a member of CC35,11 was determined to be CC20 in this study (Supplementary Table). Both DST 659 and DST 693 clusters were greatly expanded during 2000-2011. So far, thirty-three isolates including 29 from Taiwan and 4 from Korea, were classified by eBURST as CC20 (Table 5), suggesting that CC20 is an Asian cluster, which
constitutes MLST clade 3. Interestingly, when comparing with CC4, another clade 3 cluster, CC20 isolates showed a significant expansion in north Taiwan.
Among the 285 isolates characterized, only two fluconazole resistant isolates were detected. This result is consistent with the previous report that most C. albicans isolates causing invasive infections are susceptible to antifungal drugs.3,23-26 This low
rate of fluconazole resistance may be explained by fewer patients having prior fluconazole treatments in Taiwan.27 No significant antifungal susceptibility trend of the 9 drugs was observed during 2003-2011 (data not shown). It is worth noting that most (15/19, 78.9%) DST 693 isolates and CC20 cluster isolates showed a lower susceptibility to fluconazole. DST 693 is predominant in the CGMHL isolates. It was first discovered (MIC = 0.25 µg/ml) in Taiwan from the sputum of an AIDS patient in 1996.11,28 It is likely that isolates of DST 693 have been in existence in Taiwan for
years and have gained some anti-fluconazole activity since then. The decrease in fluconazole sensitivity may benefit DST 693 or CC20 isolates than other clade 3 isolates during prophylactic fluconazole treatment, and that probably makes CC20 expansion in north Taiwan.
Acknowledgments
We appreciate the assistance of Professor Frank C. Odds in assigning new allele 251 252 253 254 255 256 257 258 259 260 261 262 263 264 265 266 267 268 269 270 271 272 273 274 275
numbers and DSTs of the CGMHL isolates. We thank Dr. Chao-Hung Lee for editing the manuscript and Dr. Hsin-Fu Liu for helpful discussions in phylogenetic analyses. This work was supported by grants from Chang Gung Memorial Hospital
(CMRPG3B1302, CMRPG3D1241, CMRPG3D1611) in Taiwan.
Declaration of interest
All authors declare no conflicts of interest in this article. 276 277 278 279 280 281 282 283
REFERENCES
1. De Rosa FG, Trecarichi EM, Montrucchio C, et al. Mortality in patients with early- or late-onset candidaemia. J Antimicrob Chemother. 2013;68(4):927-935.
2. Ortega M, Marco F, Soriano A, et al. Candida species bloodstream infection: epidemiology and outcome in a single institution from 1991 to 2008. J Hosp
Infect. 2011;77(2):157-161.
3. Ruan SY, Hsueh PR. Invasive candidiasis: an overview from Taiwan. J
Formos Med Assoc. 2009;108(6):443-451.
4. Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev. 2007;20(1):133-163.
5. Chen LY, Kuo SC, Wu HS, et al. Associated clinical characteristics of patients with candidemia among different Candida species. J Microbiol Immunol
Infect. 2013;46(6):463-468.
6. Pyrgos V, Ratanavanich K Fau - Donegan N, Donegan N Fau - Veis J, Veis J Fau - Walsh TJ, Walsh Tj Fau - Shoham S, Shoham S. Candida bloodstream infections in hemodialysis recipients. Med Mycol. 2009;47(5):463-467. 7. Cheng MF, Yang YL, Yao TJ, et al. Risk factors for fatal candidemia caused
by Candida albicans and non-albicans Candida species. BMC Infect Dis. 2005;5:22.
8. Sheridan RL, Weber JM, Budkevich LG, Tompkins RG. Candidemia in the pediatric patient with burns. J Burn Care Rehabil. 1995;16(4):440-443. 9. Lortholary O, Dupont B. Antifungal prophylaxis during neutropenia and
immunodeficiency. Clin Microbiol Rev. 1997;10(3):477-504.
10. McManus BA, Coleman DC. Molecular epidemiology, phylogeny and evolution of Candida albicans. Infect Genet Evol. 2014;21:166-178.
11. Odds FC, Bougnoux ME, Shaw DJ, et al. Molecular phylogenetics of Candida
albicans. Eukaryot Cell. 2007;6(6):1041-1052.
12. Bougnoux ME, Morand S, d'Enfert C. Usefulness of multilocus sequence typing for characterization of clinical Isolates of Candida albicans. J Clin
Microbiol. 2002;40(4):1290-1297.
13. Bougnoux ME, Aanensen DM, Morand S, Theraud M, Spratt BG, d'Enfert C. Multilocus sequence typing of Candida albicans: strategies, data exchange and applications. Infect Genet Evol. 2004;4(3):243-252.
14. Gong YB, Zheng JL, Jin B, et al. Particular Candida albicans strains in the digestive tract of dyspeptic patients, identified by multilocus sequence typing.
PLoS One. 2012;7(4):e35311.
284 285 286 287 288 289 290 291 292 293 294 295 296 297 298 299 300 301 302 303 304 305 306 307 308 309 310 311 312 313 314 315 316 317 318 319 320
15. Tavanti A, Davidson AD, Fordyce MJ, Gow NA, Maiden MC, Odds FC. Population structure and properties of Candida albicans, as determined by multilocus sequence typing. J Clin Microbiol. 2005;43(11):5601-5613. 16. MacCallum DM, Castillo L, Nather K, et al. Property differences among the
four major Candida albicans strain clades. Eukaryot Cell. 2009;8(3):373-387. 17. Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring
patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186(5):1518-1530. 18. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular
Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725-2729.
19. Pfaller MA, Diekema DJ. Progress in antifungal susceptibility testing of
Candida spp. by use of Clinical and Laboratory Standards Institute broth
microdilution methods, 2010 to 2012. J Clin Microbiol. 2012;50(9):2846-2856.
20. Pfaller MA, Rhomberg PR, Messer SA, Jones RN, Castanheira M.
Isavuconazole, micafungin, and 8 comparator antifungal agents' susceptibility profiles for common and uncommon opportunistic fungi collected in 2013: temporal analysis of antifungal drug resistance using CLSI species-specific clinical breakpoints and proposed epidemiological cutoff values. Diagn
Microbiol Infect Dis. 2015:In press.
21. Bougnoux ME, Tavanti A, Bouchier C, et al. Collaborative consensus for optimized multilocus sequence typing of Candida albicans. J Clin Microbiol. 2003;41(11):5265-5266.
22. Odds FC. Molecular phylogenetics and epidemiology of Candida albicans.
Future Microbiol. 2010;5(1):67-79.
23. Pfaller MA, Castanheira M, Messer SA, Jones RN. In vitro antifungal
susceptibilities of isolates of Candida spp. and Aspergillus spp. from China to nine systemically active antifungal agents: data from the SENTRY antifungal surveillance program, 2010 through 2012. Mycoses. 2015.
24. Bonfietti LX, Szeszs MW, Chang MR, et al. Ten-year study of species distribution and antifungal susceptibilities of Candida bloodstream isolates at a Brazilian tertiary hospital. Mycopathologia. 2012;174(5-6):389-396.
25. Yang YL, Cheng MF, Wang CW, et al. The distribution of species and susceptibility of amphotericin B and fluconazole of yeast pathogens isolated from sterile sites in Taiwan. Med Mycol. 2010;48(2):328-334.
26. Cheng MF, Yu KW, Tang RB, et al. Distribution and antifungal susceptibility of Candida species causing candidemia from 1996 to 1999. Diagn Microbiol 321 322 323 324 325 326 327 328 329 330 331 332 333 334 335 336 337 338 339 340 341 342 343 344 345 346 347 348 349 350 351 352 353 354 355 356 357 358
Infect Dis. 2004;48(1):33-37.
27. Yang YL, Cheng MF, Chang YW, et al. Host factors do not influence the colonization or infection by fluconazole resistant Candida species in hospitalized patients. J Negat Results Biomed. 2008;7:12.
28. Chen KW, Chen YC, Lo HJ, et al. Multilocus sequence typing for analyses of clonality of Candida albicans strains in Taiwan. J Clin Microbiol.
2006;44(6):2172-2178.
29. Shin JH, Bougnoux ME, d'Enfert C, et al. Genetic diversity among Korean
Candida albicans bloodstream isolates: assessment by multilocus sequence
typing and restriction endonuclease analysis of genomic DNA by use of BssHII. J Clin Microbiol. 2011;49(7):2572-2577.
359 360 361 362 363 364 365 366 367 368 369 370
Table 1. Clade distribution of isolates from CGMHL and other areas.
Clade
No. of Isolates
from CGMHL No. of Isolates (%)#
Known DST New DST CGMHL Asia† Global (after 2000) Global (before 2000) 1 29 25 54 (18.9) 53 (31)** 249 (29.9)*** 156 (44.1)*** 2 0 0 0 (0) 3 (1.8) 111 (13.3)*** 35 (9.9)*** 3 25 6 31 (10.9) 8 (4.7)* 67 (8.0) 31 (8.8) 4 37 12 49 (17.2) 15 (8.8)* 127 (15.2) 33 (9.3)* 5 2 5 7 (2.5) 4 (2.3) 12 (1.4) 6 (1.7) 6 2 2 4 (1.4) 5 (2.9) 20 (2.4) 13 (3.7) 7 1 0 1 (0.4) 1 (0.6) 7 (0.8) 7 (2.0) 8 7 8 15 (5.3) 11 (6.4) 29 (3.5) 9 (2.5) 9 5 5 10 (3.5) 7 (4.1) 35 (4.2) 8 (2.3) 10 5 6 11 (3.9) 2 (1.2) 9 (1.1)** 4 (1.1)* 11 9 2 11 (3.9) 7 (4.1) 74 (8.9)** 16 (4.5) 12 6 3 9 (3.2) 5 (2.9) 15 (1.8) 9 (2.5) 13 0 0 0 (0) 1 (0.6) 3 (0.4) 1 (0.3) 14 5 2 7 (2.5) 11 (6.4)* 13 (1.6) 1 (0.3)* 15 1 2 3 (1.1) 4 (2.3) 10 (1.2) 2 (0.6) 16 17 12 29 (10.2) 6 (3.5)* 7 (0.8)*** 2 (0.6)*** 17 16 15 31 (10.9) 14 (8.2) 23 (2.8)*** 7 (2.0)*** 18 5 5 10 (3.5) - - -Singlet 0 3 3 (1.1) 14 (8.2)*** 22 (2.6) 14 (4.0)*** Total 172 113 285 (100) 171 (100) 825 (100) 362 (100) #: Number of Asian and global isolates were reported by Odds et al..11 *: p<0.05; **:
p<0.01; ***: p<0.001.
†: More than 95% of Asia isolates were collected after year 2000. 371
372
373 374 375
Table 2. eBURST clonal distribution of C. albicans isolates. Clonal Complex Predicte d Founder No. of DSTs in CGMHL Isolates No. of Isolates (%)# CGMHL Asia†† Global† 1 69 10 15 (5.3) 26 (7.2) 600 (24.6)*** 2 124 1 1 (0.4) 10 (2.8)* 248 (10.2)*** 3 155 0 0 (0) 2 (0.6) 221 (9.1)*** 4 344 2 2 (0.7) 6 (1.7) 87 (3.6)* 5 538 0 0 (0) 0 (0) 71 (2.9)** 6 299 7 9 (3.2) 13 (3.6) 54 (2.2) 7 735 6 7 (2.5) 4 (1.1) 55 (2.3) 8 659 18 45 (15.8) 21 (5.8)*** 38 (1.6)*** 9 766 12 25 (8.8) 19 (5.3) 35 (1.4)*** 10 461 6 10 (3.5) 4 (1.1) 39 (1.6)* 11 409 4 4 (1.4) 0 (0)* 38 (1.6) 12 304 5 8 (2.8) 2 (0.6)* 23 (0.9)* 13 727 6 7 (2.5) 17 (4.7) 20 (0.8)* 14 365 2 5 (1.8) 11 (3) 35 (1.4) 15 669 12 24 (8.4) 2 (0.6)*** 8 (0.3)*** 16 90 0 0 (0) 4 (1.1) 24 (1) 17 443 10 23 (8.1) 6 (1.7)*** 9 (0.4)*** 18 840 1 1 (0.4) 2 (0.6) 17 (0.7) 19 439 4 6 (2.1) 12 (3.3) 14 (0.6)* 20 693 7 27 (9.5) 6 (1.7)*** 9 (0.4)*** Others - 65 66 (23.2) 19 4(53.7) 790 (32.4) Total 178 285 (100) 361(100) 2435(100) #: *: p<0.05; **: p<0.01; ***: p<0.001.
†: Global isolates includes all isolates in the MLST database (date accessed 10.01.14). ††: Asian isolates include those from China, Hong Kong, Japan, Korea/South Korea,
Malaysia, and Taiwan recorded in the MLST database (date accessed 10.01.14) . 376 377 378 379 380 381 382
Table 3. Correlation between UPGMA grouping and eBURST clustering of CGMHL isolates.
UPGMA
Clade (No. of Isolates) eBURST Clonal Complex (No. of Isolates) Clade 1 (54) CC1 (15), CC9 (25), CC50 (2), Singleton (12) Clade 2 (0) -Clade 3 (31) CC4 (2), CC20 (27), Singleton (2) Clade 4 (49) CC2 (1), CC8 (45), Singleton (3) Clade 5 (7) CC33 (1), CC41 (1), CC79 (2), Singleton (3) Clade 6 (4) CC11 (4) Clade 7 (1) CC18 (1) Clade 8 (15) CC14 (5), CC23 (1), CC32 (2), CC68 (1), Singleton (6) Clade 9 (10) CC7 (7), Singleton (3) Clade 10 (11) CC12 (8), Singleton (3) Clade 11 (11) CC10 (10), Singleton (1) Clade 12 (9) CC6 (9) Clade 13 (0) -Clade 14 (7) CC19 (6), Singleton (1) Clade 15 (3) CC25 (1), CC59 (2) Clade 16 (29) CC15 (24), Singleton (5) Clade 17 (31) CC17 (23), CC22 (1), CC31 (3), Singleton (4) Clade 18 (10) CC13 (7), Singleton (3) 383 384 385 386 387
Table 4. CGMHL C. albicans isolates with lower fluconazole susceptibility. Clonal Complex Predicted Founder No. of Isolates
No. of Isolates with MIC>=0.5 (%)# 1 69 15 5 (33.3) 6 299 9 2 (22.2) 7 735 7 3 (42.9) 8 659 45 27 (60.0) 9 766 25 15 (60.0) 10 461 10 10 (100) ** 11 409 4 4 (100) 12 304 8 4 (50.0) 13 727 7 4 (57.1) 14 365 5 0 (0) * 15 669 24 5 (20.8) ** 17 443 23 13 (56.5) 19 439 6 2 (33.3) 20 693 27 23 (85.2) *** 31 466 3 1 (33.3) Others† - 67 32 (47.8) Total - 285 150 (52.6) #: **: p<0.01; ***: p<0.001.
†: Others are the isolates belonging to singletons or those in the CCs with less than 3 isolates. 388 389 390 391 392 393 394
Table 5. DST and geographic distribution of eBURST clonal complex 20 isolates.
DST IsolateNo. of MLST ID Strain Country Reference
674 1 923 P06 Taiwan Ref.28 693 (clonal founder) 20 944 P20-1 Taiwan Ref.28 CGMHL C006, C040, C049, C070, C072, C105, C109, C122, C163, D027, D038, D048, P004, P020, P043, P059, P061, P067, P068
Taiwan This Study
1563 3 1872 C6355 South Korea
MLST database
CGMHL C066, C082 Taiwan This Study
1564 2 1873 C4502 South Korea
MLST database
CGMHL C141 Taiwan This Study
1796 3 2019 C1098 Korea
MLST database
CGMHL C031, C096 Taiwan This Study
2006 1 2115 NICU_6643 Korea databaseMLST
2433 1 CGMHL C022 Taiwan This Study
2437 1 CGMHL C038 Taiwan This Study
2451 1 CGMHL C085 Taiwan This Study
395 396
397 398
Figure Legends
Figure 1. MLST analysis of CGMHL C. albicans isolates. Nucleotide sequences of 7 housekeeping genes of 285 CGMHL isolates and 996 reference strains retrieved from the MLST database were analyzed by UPGMA. Clade numbers were assigned as described previously.11,29 Clades 2 and 13 (in
italic letters) contained no CGMHL isolates. Open circles represent CGMHL isolates. The scale bar indicates p-distance. 399 400 401 402 403 404