國立 交 通 大 學
生物科技研究所
碩 士 論 文
黑腹果蠅蛋白質酪氨酸亞硫酸化酵素之鑑定與
分析
Identification and Characterization of the Enzyme
Responsible for Protein Tyrosine Sulfation in Drosophila
melanogaster
研究生: 王晨竹
指導教授: 楊裕雄 教授
中華民國九十九年七月
黑腹果蠅蛋白質酪氨酸亞硫酸化酵素之鑑定與分析
Identification and Characterization of the Enzyme
Responsible for Protein Tyrosine Sulfation in Drosophila
melanogaster
研 究 生:王晨竹 Student: Chen-Chu Wang
指導教授:楊裕雄 教授 Advisor: Prof. Yuh-Shyong Yang
國立交通大學
生物科技學系
碩 士 論 文
A Thesis
Submitted to Department of Biological Science and
Technology
National Chiao Tung University
in Fulfillment of the Requirements
for the Degree of
Master of Science
in
Biologic Science and Technology
July 2010
Hsinchu, Taiwan, Republic of China
i
黑腹果蠅蛋白質酪氨酸亞硫酸化酵素之鑑定與分析
學生: 王晨竹
指導教授: 楊裕雄 教授
國立交通大學生物科技學系碩士班
摘要
蛋白質酪氨酸的亞硫酸化為許多特定分泌性蛋白質或膜蛋白質很重
要的後修飾。酪氨酸亞硫酸基轉移酶(tyrosylprotein sulfotransferase)
負責的酪氨酸亞硫酸化,在細胞間的蛋白質與蛋白質交互作用,和許
多重要的生物功能反應擔任關鍵的調節作用,其中包含人類免疫缺陷
病毒(HIV)感染入侵、發炎反應、凝血機制、不孕...等等。然而這些
生理與病理的機制都還是不清楚的。利用已經被解碼的染色體序列和
生物資訊系統分析的協助,搜尋表達序列標記(EST)的資料庫,發現
有一段基因可能負責黑腹果蠅體內酪氨酸亞硫酸化的反應。本研究將
具有酵素活性的黑腹果蠅酪氨酸亞硫酸基轉移酶(DmTPST)表達在大
腸桿菌BL21(DE3)pLysS寄主细胞,純化出高產率均質的酵素,並且
探討其特性;利用polyEAY當作受質,定義出黑腹果蠅酪氨酸亞硫酸
基轉移酶理想的最佳反應狀況。最後,藉由基因重組的酪氨酸亞硫酸
基轉移酶催化了drosulfokinin的亞硫酸化,證明能夠進一步的解釋黑
腹果蠅蛋白質酪氨酸的亞硫酸化未知的機制與功能。
ii
Identification and Characterization of the Enzyme Responsible for
Protein Tyrosine Sulfation in Drosophila melanogaster
Student: Chen-Chu Wang Advisor: Prof. Yuh-Shyong Yang Department of Biological Science and Tenology and Institute of Biochemical
Engineering, National Chiao Tung University, Hsinchu, Taiwan, ROC
ABSTRACT
Protein tyrosine sulfation, catalyzed by tyrosylprotein sulfotransferase (TPST), is one of the most common post-translational modifications towards secretory and transmembrane proteins. Protein tyrosine sulfation is a key modulator of extracellular protein-protein interactions and responsible for various important biological functions including HIV entry, inflammation, coagulation, and sterility. These physiological and pathological mechanisms, however, are not clear. With the assistances of decoded genome sequences and bioinformatic analysis, a promising gene competent for catalysis of protein tyrosine sulfation in Drosophila melanogaster was discovered by searching the expressed sequence tag (EST) database. Enzymatically active
Drosophila melanogaster TPST (DmTPST) was first cloned, expressed in Escherichia coli BL21(DE3)pLysS host cells and purified to homogeneity in high yield. The
homogeneous DmTPST was characterized through radioactive assay with polyEAY as substrate and its optimal reaction conditions were determined. Finally, the drosulfokinin sulfation catalyzed by recombinant DmTPST was firstly demonstrated, which provided direct link to tyrosine sulfation in Drosophila melanogaster and further opportunity to decipher the obscure mechanisms and functions of protein sulfation.
iii
Acknowledgement
不知不覺的碩士班就過完了。當初剛考來交大時,還很擔心碩士的生活有沒有辦 法很順利,畢竟來到了一個新的環境,而且碩士生活不像大學生一樣只需要拼命 唸書就行,所以深怕無法步入軌道,但是在楊裕雄老師的實驗室中,從一剛開始 的新生訓練開始,一步一步的將我帶入了研究的領域當中,在我碩士研究的期間, 要感謝的人實在好多,真是多虧了這些夥伴在精神上或是實質上的幫助,才讓我 順利的拿到碩士畢業證書。首先要感謝的就是我的老闆楊裕雄教授,看到老師總 是帶著笑容亦師亦友的跟我們分享許多實驗或生活中的經驗,並且提供我們一個 良好的研究環境,開啟我的研究生涯並且讓我了解生為一個研究生應該具備的觀 念。再來是已經畢業離開 LEPE 實驗室的學長姐們:小米學姊,謝謝妳當初在實 驗上的教導,讓我了解到實驗操作中嚴謹的態度。秀華學姊,坐在我隔壁,跟我 說了實驗室的一些守則跟大家的個性,讓我融入其中。音汝學姊,當初要出國時 妳就像是導遊一樣,規劃的非常周到,讓我們可以好好參加研討會又可以觀光。 再來是生電組的組員:呈允學長,一直要到了你快畢業時的這幾個月才跟你比較 熟,歡迎你以後時常回來跟我們敘敘舊。淵仁學長,一直耳聞你興趣非常廣泛, 能文能武,以會有機會再跟你請教。小志學長,非常謝謝你時常在 meeting 時從 不同的觀點給我不同的意見,讓我了解我報告中的缺失。俊瑋,你非常的厲害, 學習能力很強,但你現在正在休學中,也期待你可以回來,繼續你未完成的學業。 欣怡,也恭喜你要跟我們一起畢業囉,以後也歡迎回來敘舊。曉萍,未來生電組 的棟樑,生電組的一姊就由妳接手,把生電組帶向另一個境界吧。還有介於生電 組與酵素組之間的第三類組,就成為整合組吧,這組唯一的成員奇叡,接下來你 可要好好的我們兩組的研究架起一道橋樑吧。接下來就是我們酵素組的成員:首iv 先是我這組的組長,陸宜學長,非常感謝你在我們這組的貢獻,把我們這組照顧 的這麼好,你不管在研究或是各方面都是略懂一點,但在我看來似乎只是謙虛而 已,在接下來的日子裡,我還有很多要跟你學習的地方,希望你能夠多多包含囉。 普普學長,也是一位非常有趣的學長,非常感謝你在實驗上給的意見,我也覺得 你懂的東西非常多,之後也希望你能夠多多的教導。小胖學長,謝謝你當初帶我 們做新生訓練,其實我知道你都是為新生好,所以才會比較嚴苛,但這一切都是 等到我跟你比較熟後才體會的,有你在我們這組增加了許多的樂趣,往後也請你 多多照顧,一起弄好酵素組吧。咏馨,從學姊變成同學,第一次看到妳時,就覺 得妳很厲害,畢竟也在 LEPE 待久了,所以很多實驗室的狀況妳都知道,接下來 也要一起好好的把酵素組的研究給延伸下去。資翔,也恭喜你要跟我們一起畢業 了,也非常感謝你延伸我的研究,雖然你沒有時間可以完成後續發表的工作,但 我好好好的整合你的研究,到時一起發表出去。芝綺,身為酵素組,但做的研究 也是有跨到整合的部份,未來也希望妳能夠好好的把我們這組的研究給應用到生 電組中吧。還有 Sonia,很感謝妳幫我們處理實驗室帳務上的問題,讓我們省去 許多的時間。接下來就是我的同袍們:文燦與康寧,總是跟我在一起喇賽,雖然 沒在同一組,但是你們總是可以給我一些實驗上的意見,而且也謝謝你們幫忙我 處理實驗室的一些大小事,接下來也希望文燦可以找到好工作,當個傑出學長贊 助 LEPE;康寧可以趕緊畢業,也期待妳健身有成囉。而且這一年來,實驗室的 氣氛越來越好,實在是很喜歡這種感覺,希望以後可以一直維持下去,畢業的各 位,也多多回來關照與敘舊,讓新生們了解你們的豐功偉業。接下來要感謝各位 參加我碩士論文口試的老師們:彭惠玲、汪宏達、簡慶德老師們,感謝你們再我 碩士口試中給的意見,讓我知道當中的缺失,讓我可以好好的修改我的碩士論文,
v 使論文更加的完整。最後要感謝的就是我的家人:爸爸跟媽媽,總是擔心我們學 業狀況,也擔心未來的出入,也給我許多自主權,讓我沒有經濟的壓力,可以順 利的唸完碩士。阿公跟阿嬤,年紀都很大了,寧願省吃儉用也要給我們這些孫子 們好的照顧。姊姊跟妹妹,也很謝謝妳們給我的關心與照顧囉。致謝就寫到這邊, 希望曾經幫助過我的大家都可以平安、順心,謝謝!
vi
Contents
Chinese abstract ... i English abstract ... ii Acknowledgement ... iii Contents ... viContents of Tables ... viii
Contents of Figures ... ix Contents of Appendices ... x Abbreviations ... xi 1. Introduction ... 1 1.1 Post-translational modifications ... 1 1.2 Sulfotransferase ... 1 1.3 Tyrosylprotein sulfotransferase ... 3
1.4 Biological functions of protein tyrosine sulfation ... 4
1.4.1 Chemokine receptor ... 4
1.4.2 Leukocyte adhesion and inflammatory response ... 5
1.4.3 Hemostasis and anticoagulation ... 6
1.5 Bottlenecks of protein sulfation research ... 7
1.6 Tyrosylprotein sulfotransferase in Drosophila melanogaster ... 8
1.7 Contribution from this sutdy ... 9
2. Materials ... 10
3. Experimental procedures ... 11
3.1 Prediction of transmembrane domain of DmTPST ... 11
3.2 Cloning of DmTPST... 11
vii
3.4 Mass analysis ... 12
3.5 A DmTPST enzymatic activity assay ... 12
3.6 Using enzymatic activity assay characterize DmTPST ... 13
4. Results ………... 15
4.1 Expression of recombinant Drosophila melanogaster TPST in prokaryote expression system ... 15
4.2 Sulfation of polyEAY in a PAPSS and TPST coupled system ... 15
4.3 Determination of DmTPST effective range and time course of DmTPST catalysis ... 16
4.4 pH profile of DmTPST ... 16
4.5 Kinetics of DmTPST utilized polyEAY as substrate ... 17
4.6 DmTPST sulfation reaction in drosulfakinins ... 17
5. Discussion... 18
References ... 23
Tables ... 30
Figures ... 32
viii
Contents of Tables
Table.1 Purification of NusA-DmTPST from E. coli... 30 Table.2 Coupled enzyme assay-obtained kinetic characterization of DmTPST in comparison with previous radiometric assay ... 31
ix
Contents of Figures
Figure.1 Scheme for the determination of TPST activity ... 32
Figure.2 Bioinformation analyze protein sequence identity and transmembrane domain for human and Drosophila melanogaster ... 33
Figure.3 Purification homogenous recombinant DmTPST ... 34
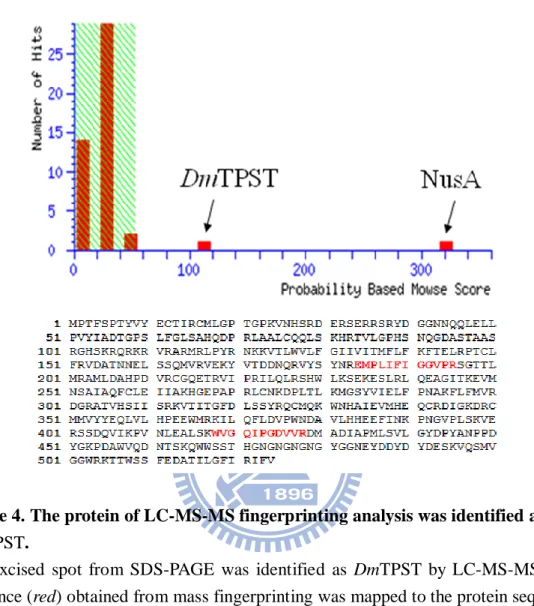
Figure.4 The protein of LC-MS-MS fingerprinting analysis was identified for Drosophila melanogaster TPST ... 35
Figure.5 Autoradiography of [35S]-labeled polyEAY catalyzed by DmTPST .... 36
Figure.6 Effective range of DmTPST assay ... 37
Figure.7 Time course effects on the activity of recombinant DmTPST ... 38
Figure.8 pH profile effects on the activity of recombinant DmTPST ... 39
Figure.9 Kinetics of DmTPST using polyEAY as substrate ... 40
Figure.10 Autoradiography of [35S]-labeled Drosulfakinin catalyzed by DmTPST . ... 41
x
Contents of Appendices
Appendix 1. Schematic representation of protein modifications related to the
regulation of biological processes ... 42
Appendix 2. Some common and important post-translation modifications ... 43
Appendix 3. General ST-catalyzed reaction with PAPS as the cosubstrate ... 44
Appendix 4. Sulfate activation and tyrosine O-sulfation ... 45
Appendix 5. Schematic representation of cell entry by HIV-1 following sulfonation of CCR5 by a tyrosylprotein sulfotransferase ... 46
Appendix 6. Tyrosine sulfation plays an important role in the immune response ... 47
Appendix 7. pET-43.1a-c(+) Vector exposition... 48
Appendix 8. MS analysis of DmTPST... 49
xi
Abbreviations
Abbreviation and Symbol Full name
Absorption (exitinction) coefficient
A280 Absorption at 280 nm
A600 Absorption at 600 nm
CCK Cholecystokinin
CCR5 Chemokine (C-C motif) receptor 5 CD4 Cluster of differentiation 4
D. melanogaster Drosophila melanogaster
gp120 Glycoprotein 120
HIV Human immunodeficiency virus
kcat Turnover number
kDa Kilodaton
Km Michaelis constant
MALDI-TOFt Matrix-assisted laser desorption ionization time of flight MES 2-[N-morpholino] ethanesulfonic acid
PAGE Polyacrylamide gel electrophoresis
PAP Adenosine 3',5'-diphosphate
PAPS 3'-phosphoadenosine 5'-phosphosulfate polyEAY polypeptide(Glu : Ala : Tyr = 6 : 3 : 1) PSGL-1 P-selectin glycoprotein ligand-1 PTM Post-translational modification
SDS Sodium dodecyl sulfate
STs Sulfotransferases
TPST Tyrosylprotein sulfotransferase
1
Introduction
1.1 Post-translational modifications
Post-translational modifications (PTMs) are one of the most important
biological in both prokaryote and eukaryote proteins that can regulate the protein
functions and activities by causing the changes of the protein structure or the affinity
of dynamic interaction between proteins and compounds. (Appendix 1) (Seo et al.
2004) Some common and important post-translational modifications include
acetylation, acylation, glycosylation, methylation, phosphorylation, ubiquitination,
and sulfation (Appendix 2). These modifications can have both structural and
regulatory functions, which modulate the properties of proteins by proteolytic
cleavage or by the addition of a modifying group to amino acid, which may involve proteins’ activity state, localization, turnover, and interaction with other proteins. Sulfation and phosphorylation are similar in modifying group, mass altered, and
molecular interaction. (Mann et al., 2003).
1.2 Sulfotransferases
Sulfonation reactions are usually classified by the acceptorgroup involved in
sulfoconjugation, for instance, O-sulfonation (ester), N-sulfonation (amide), and
S-sulfonation (thioester). O-Sulfonation is dominant in cellular sulfation reaction
2
endogenous compounds such as catecholamines, steroids, thyroid hormones, and
vitamins, and macromolecules such as glycosaminoglycans, proteoglycans, proteins,
and galactoglycerolipids. N-Sulfonation is a crucial reaction in the modification of
carbohydrate chains in macromolecules such as heparin, heparan sulfate
proteoglycans, and also involved in the metabolism of xenobiotics. (Strott, 2002).
Sulfate-containing biomolecules were identified in 1876 (Baumann et al. 1876), but the mechanism of sulfation remains unknown until the active 3’-phosphoadenosine 5’-phosphosulfated (PAPS) was isolated. Sulfotransferases(STs) use PAPS as sulfate group (SO3-) donor, to catalyze the sulfuryl group into a variety of amine and
hydroxyl substrates (Appendix 3). STs can be basically divided into two classes:
cytosolic STs and membrane-associated STs. Cytosolic STs are soluble proteins
located in cytoplasm, and mediated small chemical compounds including steroids,
xenobiotics, dietary carcinogens, and neurotransmitters. They are involved in
detoxification, hormone regulation, and drug metabolism. Membrane-associated STs
are membrane anchored proteins located in the trans-Golgi network (TGN), which
implied that they are involved in the post-translational modification of larger
biomolecules including carbohydrates and protein such as heparan, glycoproteins, and
oligopeptide. They are mainly involved in molecular-recognition events and
3
1.3 Tyrosylprotein sulfotranferases
Tyrosine O-sulfation of protein was first discovered in bovine fibrinopeptide B
by Bettelheim in 1954 (Bettelheim, 1954). However, limited information was known
about tyrosylprotein sulfation until 1982, when Huttner directly identified that this
PTM was mediated by tyrosylprotein sulfotransferase (TPST), an enzyme that
catalyzes the transfer of a sulfuryl group from PAPS to the hydroxyl group of tyrosine
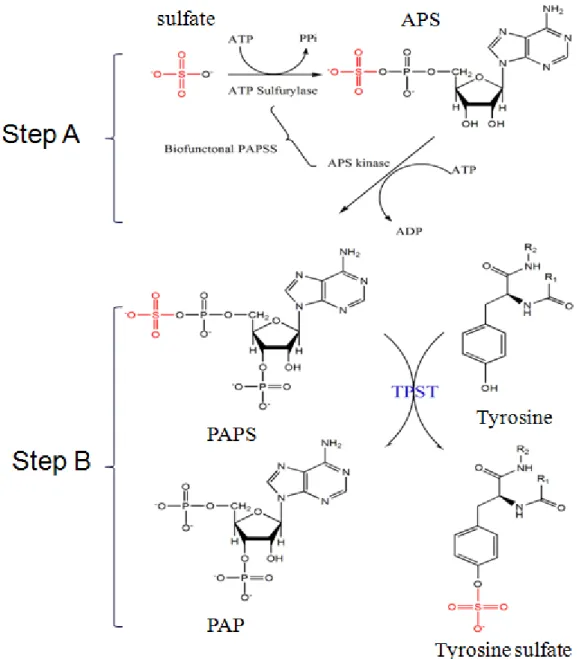
residue in the protein/peptide (Fig. 1 step B) (Moore, 2003). Furthermore, Huttner
proved that TPST was membrane-bound and located in the trans-Golgi network
(Appendix 4) (Baeuerle and Huttner, 1987), and also characterized and purified
TPST from bovine adrenal medulla (Niehrs and Huttner, 1990). It is now known that
TPST is a widespread enzyme in multicellular eukaryotic organisms throughout the
plant and animal kingdoms, and can be detected in most tissues and cell types from
humans and rats.(Mishiro et al., 2006) (Nishimura and Naito, 2007). TPST are type II
transmembrane proteins with a short N-terminal cytoplasmic domain, a single about
17-residue transmembrane domain (red), and a luminal catalytic domain.(Fig.
2)(Baeuerle et al. 1987; Lee et al. 1985). In most species, TPST have two isoenzyme: TPST-1 and TPST-2, but D. melanogaster have only a single TPST gene. The
TPST-1 and TPST-2 share 65-68% sequence identity. Furthermore,
tyrosine-sulfated proteins, TPST activity or putative TPST orthologs have not been
4
developed a software, called Sulfinator (http://ca.expasy.org/tools/sulfinator)
(Monigatti et al., 2002), which predicts possible proteins that can process tyrosine
sulfation and also its tyrosine sulfation site. It has been estimated that up to
approximately 1% of all tyrosine residues in eukaryotic cells are predicted to undergo
tyrosine sulfation, but only a few hundred proteins have been identify presently
(Seibert and Sakmar, 2008).
1.4 Biological functions of protein tyrosine sulfation
TPSTs catalyze the sulfation of tyrosine residues within specific peptide
sequences, which have also been implicated in several crucial physiological and
pathological mechanisms. Current thinking holds that this PTM serves as a key
modulator of protein–protein interactions of secreted and membrane-bound proteins
(John W Kehoe et al., 2000). Tyrosine sulfation has been implicated in intracellular
trafficking and proteolytic processing of secreted proteins, and a key modulator of
extracellular protein-protein interactions, which includes hormonal regulation,
hemostasis, inflammation and infectious diseases (Seibert and Sakmar, 2008).
1.4.1 Chemokine receptor
5
through G-protein-coupled receptors, including leukocyte trafficking, angiogenesis,
angiostasis, viral infections, and host immune response to cancer (Zlotnik et al., 1999).
Several chemokine receptors (CXCR3, CXCR4, CCR2b, CCR5, and CX3CR1) have
been shown to undergo tyrosine sulfation (Farzan et al. 1999; Farzan et al. 2002; Preobrazhensky et al. 2000; Fong et al. 2002; Colvin et al. 2006). Currently, the most popular topic on the study of tyrosine sulfation focuses on CCR5 due to its
involvement of HIV-1 entry. The chemokine receptor CCR5 is post-translationally
modified by sulfation of its N-terminal tyrosines.Sulfated tyrosines contribute to the
binding of MIP-1α, MIP-1β, and HIV-1 gp120/CD4 complexes and to facilitator
HIV-1 to enter cells expressing CCR5 and CD4 (Appendix 5). The N terminus of
CCR5 contains four tyrosines at positions 3, 10, 14, and 15(Samson et al., 1996), and
modified stepwise at positions 14 or 15, followed by position 10 and finally the
tyrosine residue at position 3 (Sasaki et al., 2007). Mutation of the four sulfotyrosine
residues in CCR5 to phenylalanine and chlorate inhibits HIV infection by 50–75%.
This information suggests that inhibiting tyrosine sulfation of CCR5 may provide a
basis for the design of therapeutic agents aimed at blocking HIV-1 cellular entry.
1.4.2 Leukocyte adhesion and inflammatory response
6
cell and endothelial cells that binds to P-selectin. In immune response, the leukocytes
need to reach the inflammation site through passage of the blood circulation, the then
roll upon, adhere to, and finally transmigrate between the endothelial cells and
infective site (Appendix 6a). The extreme amino terminus of PSGL-1 carries three
tyrosine sulfation sites. These sulfate esters, and specific glycans on PSGL-1, are key
binding determinants for P-selectin (Appendix 6b). The binding between PSGL-1 of
leukocyte and P-selectin of endothelial cells is essential for leukocyte adhesion in this
inflammatory response (Kehoe and Bertozzi, 2000; Pouyani and Seed, 1995).
Treatment of PSGL-1 with a bacterial arylsulfatase releases sulfate from tyrosine
reduced the binding ability to P-selectin (Wilkins et al., 1995), and the results were
also supported by point mutagenesis of tyrosine (Sako et al., 1995). According to
result, TPST has become a therapeutic target for autoimmune diseases caused by
chronic inflammation, such as rheumatoid arthritis and multiple sclerosis (Hsu et al.,
2005).
1.4.3 Hemostasis and anticoagulation
The biological function of tyrosine sulfation is also involved in hemostasis.
This modification is crucial in the interaction between many plasma proteins such as
hirudin and thrombin (Stone et al. 1986), fibronectin and fibrin (Suiko et al. 1988),
7
glycoprotein (GP) Ibα with both vWF and thrombin (Marchese et al. 1995; Ward et al.
1996; Fredrickson et al. 1998; Dong et al. 2001; Murata et al. 1991). Moreover, the
complete mechanism of platelet attachment is accomplished by vWF that bridges subendothelial collagen and platelet membrane protein GP Ibα. The binding between vWF and n GP Ibα is dependent upon the sulfation of three tyrosine residues (Tyr276, 278, 279). In anticoagulation, hirudin is a potent anticoagulant protein secreted in the
salivary gland of the leech. When Tyr63 has been sulfated, the tyrosine sulfation of
hirudin has a 10-fold higher affinity for thrombin than unsulfated form, which
prevents coagulation by inhibit thrombin (Stone and Hofsteenge, 1986).
1.5 Bottlenecks of protein sulfation research
In the last five decades of studies on this topic, many questions remain
unknown about TPSTs and protein sulfation. The bottlenecks of studying TPSTs
include the difficulty of characterizing TPST due to the lack of source of homogenous
protein samples. It is hard to develop a fast and accurate assay for quantitative
kinetics analysis, because sulfation detect limit in pico-mole level. Moreover, tyrosine
O-sulfate may instability on the tyrosine residue of TPST substrate, which makes it
difficult to detect or isolate sulfated proteins and peptides. Previous research on
protein sulfation had focused on few TPST substrates as described above, therefore the understandings of TPST’s roles are restricted by the biological regulations and
8
pathways of those few substrates. Because of tools and methods are undeveloped in
protein sulfation, limited information available.
1.6 Tyrosylprotein sulfotransferase in Drosophila melanogaster
Most vertebrates (such as rat, cow, chicken, zebrafish, African clawed frog) and
invertebrates (such as Anopheles gambiae (mosquito), and Caenorhabditis elegans)
have two TPSTs. It is interesting to note that Drosophila melanogaster is so far the
only species that was discovered to contain a single TPST gene (Moore, 2003).
Therefore, D. melanogaster is a good model to study TPST, which a complete
elimination of protein sulfation modification can be reached by a simple knockout or
knockdown of a single gene. About 75% of known human disease genes have a
recognizable match in the genetic code of fruit flies (Reiter et al., 2001), and 50% of
fly protein sequences have mammalian analogues. Many advantages of using D.
melanogaster as a study model include the short generation time and easy growth.
Therefore, D. melanogaster is a suitable model to study protein tyrosine sulfation by
using complete genetic tools to understand physiological and pathological
mechanisms. The completion of genomic database is helpful for protein identification
and its function. According to speculated that there are approximately up to 1%
9
published less substrate, such as Drosulfakinin, Vitellogenin, and Glutactin (Nichols
R. et al. 1988; Baeuerle P.A. et al. 1985; Olson P.F. et al. 1990).
1.7 Contribution from this study
This was the first research focused on the identification, cloning, expression
and characterization of DmTPST at protein level. Following the expression of
DmTPST in a prokaryotic system, the desired tyrosine sulfated proteins were further
produced in vitro in a PAPS generating system. The homogeneous DmTPST was
characterized through the PAPS generating system with polyEAY as a substrate.
Large quantity of homogeneous DmTPST was obtained that facilitated further studies
and applications in protein tyrosine sulfation. Optimal reaction conditions for
DmTPST catalysis and pH profile were determined. Finally, an endogenous
compound of D. melanogaster, drosulfakinin, was demonstrated to serve as substrate
of recombinant DmTPST. The results indicated that recombinant DmTPST can further
decipher the ignorant mechanisms and functions of protein tyrosine sulfation in
10
2. Materials
Adenosine 5’-triphosphate (ATP), tris[hydroxymethyl]aminomethane (Tris), 2-[N-morpholino]ethanesulfonic acid (MES), poly-(Glu6, Ala3, Tyr1) (EAY: Mr
33KDa), inorganic pyrophosphatase, and imidazole were purchased from Sigma (St.
Louis, MO, USA). Potassium phosphate (dibasic), glycine, and sodium dodecyl
sulfate (SDS) were obtained from J. T. Baker (Phillipsburg, NJ, USA). Sodium
[35S]sulfate (1050-1600 Ci/mmol) of 99.0% radiochemical purity was purchased from
PerkinElmer (Boston, MA, USA). Taq polymerase, T4 DNA ligase, and reagents for
PCR were obtained from New England Biolabs (Beverly, MA, USA). Pfu DNA
polymerase was purchased from Stratagene (La Jolla, CA, USA). Expression vector
and BL21(DE3) pLysS competent cells were purchased from Novagen (Madison, WI,
USA). HisTrap sepharose was obtained from GE Healthcare (Uppsala, Sweden).
Cellulose thin-layer chromatography (TLC) plates were products of Merck
(Whitehouse Station, NJ, USA). All other chemicals were of the highest purity
11
3. Experimental procedures
3.1 Prediction of transmembrane domain of DmTPST
The transmembrane region and orientation of DmTPST were predicted on the
website—PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/psiform.html) (Jones, 2007).
Only scores of hydrophobicity above 0 were considered significantly to be the
potential transmembrane region.
3.2 Cloning of DmTPST
The Drosophila melanogaster TPST cDNA was subcloned into pET-43a
vector. The potential catalytic domain of TPSTs predicted above was amplified by
PCR with specific primers designed to contain BamHI in the forward direction
(5’-TGAAGAATTCGACGCCCCCAACGAGCTCTCCTC-3’) and EcoR1 restriction
sites in the reverse one contain XhoI restriction
(5’-TGCCCTCGAGCTCTCCCACAGCATTCGATTGGC-3’). cDNA fragments
were inserted into the EcoRI/XhoI double-restriction sites and then confirmed using
an ABI Prism, 346 DNA sequencer (Applied Biosystems, Foster City, CA) following
12
3.3 Protein expression and purification of DmTPST
A single colony of BL21(DE3)pLysS consisted of DmTPST plasmid was used
to inoculate in the LB broth with ampicillin as the antibiotic at 37°C. Growth was
continued to an ODA600 of 0.4–0.6 and then induced with 1 mM isopropyl-thio-β
-D-galactoside (IPTG) for 24-hr incubation at 20°C. The cultures were centrifuged at
14000g for 20 minutes, and the pellet was sonicated in IMAC5 buffer (50 mM
Tris-HCl, pH 8.0, 500 mM NaCl, 5 mM imidazole, and 10% glycerol) for DmTPST.
Further the HisTrap sepharose charged with NiSO4 was utilized to the DmTPST
purification. The homogeneous proteins were determined by SDS-polyacrylamide gel
electrophoresis (Laemmli, 1970).
3.4 Mass analysis
The in-gel digestion of interested proteins was performed by the conventional
protocol. MALDI-TOF was carried out to study the identification of excised proteins.
The PMF data was analyzed by MASCOT based on the NCBInr database.
3.5 A assay of DmTPST enzymatic activity
For the determination of DmTPST activity from bacterial expression, we
13
substrate, such as polyEAY. The couple-enzyme (human PAPS synthetase 1 and
DmTPST) radioactive assay was newly established for the measurement of DmTPST
activity. The complete assay mixture contained the following components: sulfate
acceptor DmTPST substrate (polyEAY), 4 mM inorganic Na2[35S]SO4, 5 mM
2-mercaptoethanol, 1 mM MgCl2, 50mM MES (pH6.5), 5 g recombinant human
PAPS synthetase 1 (hPAPSS1), DmTPST, and 0.5 U pyrophosphatease in a final
volume of 20 l. Assays were initiated by the addition of the hPAPSS1 and incubated
for 15 min at 37°C followed by the addition of DmTPST incubation for 45 min at
37°C. The reactions were terminated by heating at 95°C for 2 min. The supernatant
was collected and analyzed by spotting 1 l aliquot of the reaction mixture on a
cellulose thin-layer chromatography (TLC) plate and developed with
n-butanol/pyridine/formic acid/water (5:4:1:3; by volume) as the solvent system. The
dried plate was exposed with Kodak BioMax MR film which provide the optimal
resolution for 35S autoradiography.
3.6 Using enzymatic activity assay characterize DmTPST
According to TPST enzymatic activity assay control DmTPST amounts (from
s
14
DmTPST enzymatic activity of different substrates was changed from polyEAY to
15
4. Results
4.1 Expression of recombinant D. melanogaster TPST in prokaryote expression system.
The expression vector, pET-43a, harboring DmTPST cDNA was competent to
express recombinant TPST in E. coli. The prokaryotic expression of TPST was
optimized to reach the maximal soluble amount and purified to nearly homogeneity
(Fig. 3). A 96-kDa protein showed on the SDS-PAGE was composed of NusA-tag
fusion protein (60 kDa) and DmTPST (36 kDa) upon treatment in coomassie blue
R250. The spot excised from SDS-PAGE was analyzed by LC-MS/MS (Fig. 4). Two
peptides (colored in red) come after trypsin digestion the alignment of these peptide
sequences showed homology to DmTPST with high scores of confidences. The
purification table revealed the purification-related information of DmTPST (Table.
1).
4.2 Sulfation of polyEAY using a PAPSS and TPST coupled system
Autoradiography on the cellulose TLC plate demonstrated that the DmTPST
activity could be determined under the enzymatic activity assay condition (Fig. 5).
[35S]-labeled EAY produced only in the presence of DmTPST and complete PAPS
16
absence of PAPSS (lane 1), polyEAY (lane 2) or DmTPST (lane 3), respectively.
These results indicated that PAPS produced through PAPSS catalytic reactions could
be used for the sulfation of polyEAY catalyzed by DmTPST.
4.3 Determination of the effective range and time course of DmTPST enzyme catalysis
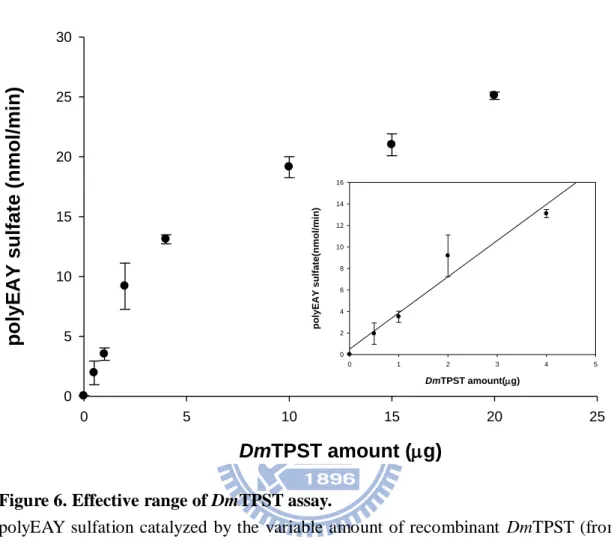
The linearly effective range of DmTPST amount in the standard assay ranged
within 5 g as shown in Fig. 6. Accordingly, 4 g DmTPST was used in further
experiments as standard assay. The time dependence of DmTPST activity with
polyEAY as substrate was examined. The concentrations of sulfate and polyEAY,
were both saturated in the reactions. The tyrosine O-sulfation of polyEAY increased
linearly with the incubation time from 15 to 120 minutes as shown in Fig. 7.
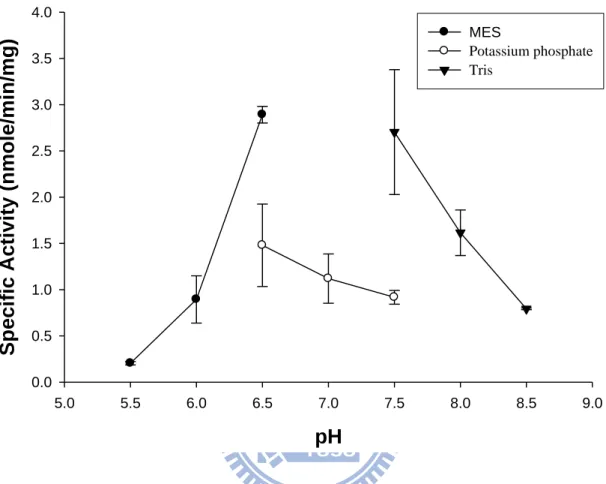
4.4 pH profile of DmTPST
pH affects the electricity of amino acid and further contributes to the substrate
binding affinity, enzymatic catalysis, and protein conformational structure. The pH
profile of the recombinant DmTPST was determined by measuring the activity at
various pH values. The pH values from 5.5 to 8.5 were shown in Fig. 8. The optimal
17
4.5 Kinetics of DmTPST with polyEAY as substrate
The kinetic constants toward polyEAY, which is synthetic polypeptides
composed of Glu, Ala, and Tyr in the ratio 6:3:1, demonstrated that Km and Vmax was
11.5 Mand 4.5 nmole/min/mg, respectively (Fig. 9). It revealed that the expression
of DmTPST was active in the catalysis and performed the similar kinetic constants
compared to the previous studies. (Table 2.)
4.6 Sulfation of endogenous substrate, drosulfakinin, by DmTPST
Drosulfakinin, composed of 14 amino acid residues, is a known endogenous substrate
in Drosophila melanogaster of DmTPST. The result from Fig. 10 revealed that the
DmTPST was competent to catalyze not only the synthetic peptide polyEAY (lane 4),
18
5. Discussion
Tyrosine sulfation was discovered in 1950s in bovine fibrinogen (Bettelheim,
1954), and afterwards, the tyrosylprotein sulfotransferase (TPST) was identified to be
responsible for this post-translational modification in 1982 (Huttner, 1982). Since the
discovery of the tyrosine O-sulfation, little about the the enzyme mechanisms have
been elucidated. This may be attributed to the lack of TPST related information, such
as the difficulty of sourcing the homogeneous enzyme and ample amount of TPST,
limited information of enzyme characteristics (kinetics), unstable sulfate groups on
the substrate, and lack of sensitive detecting methods for the sulfated tyrosine.
Drosophila melanogaster was chosen as the source of animal study due to easy
growth, short generation span, solved genomic database, well-established transgenic
tools, and more importantly, D. melanogaster only has a single TPST gene (Moore,
2003). The amino acid sequence of TPST in D.melanogaster shares 58% and 56%
with human TPST1, and TPST2, respectively (Fig. 2). Approximately 75% of known
human disease genes have a recognizable match in the genetic code of D.
melanogaster, and 50% of D. melanogaster protein sequences have mammalian
analogues (Reiter et al., 2001), which makes D. melanogaster an appropriate animal
model for pathological studies on TPST.
19
system (Lu et al., unpublished), the NusA-fused DmTPST was firstly obtained with
maximal solubility and high purity (Table 1 and lane 2 in Fig. 3), and used for
studying the enzymatic characterization. The purification yield of DmTPST showed
higher than that of hTPST2 and although the protein sequence of DmTPST and
hTPST2 has a similarity approximately 60%. The distinct characteristics between
human and D. melanogaster TPST need to study further. The NusA-DmTPST
possessed high homogeneity in our study, however, the ratio of DmTPST in this
fusion protein was merely 35% and the total molecular weight was close to 100 kDa.
The NusA protein obviously performed no interference with the enzymatic activity of
DmTPST and rendered high solubility to facilitate DmTPST folding. Overall, this
purification procedure of DmTPST was simple with stable material source, great
quantity, and homogeneous DmTPST in this study.
By the facilitation of coupled enzyme reaction (hPAPSS1 and DmTPST), the
productive rate of tyrosine sulfation was faster than that of the conventional reaction
which utilized PAPS directly as sulfate donor as shown in Fig. 1 (Liu et al.,
unpublished). The approach avoided the contamination of PAPS from PAP
(Rens-Domiano and Roth, 1989; Miller and Waechter, 1979). PAPS is extremely
costly and it tends to hydrolyze easily to form PAP, a known competitive inhibitor of
20
generated saturated PAPS from inorganic sulfate, and this scheme could obviously
prevent the background from the hydrolysis of PAPS. Moreover, the production of
protein tyrosine sulfation by this method was extremely efficient than previous studies
and it might potentially apply to spectrometric analysis in additional to radioactive
assay (Liu et al., unpublished; Mishiro et al., 2006).
In this study, DmTPST properties including the DmTPST amount (Fig. 6), time
dependence of the activities of DmTPST (Fig. 7), pH profile (Fig. 8), and kinetic
parameters of DmTPST (Fig. 9), were examined. The optimal DmTPST dosage and
reaction time was 5 g and 2 hours, respectively, which located in the linear range. In
the pH-dependent experiment, DmTPST displayed an optimal activity at pH 6.5 (Fig.
8), which was similar to that of TPST in human liver and rat submandibular salivary
glands (Lin and Roth, 1990; William et al., 1997). The result of Fig. 8 also indicated
that potassium phosphate was inhibitory to the DmTPST catalyzed sulfation of
polyEAY.
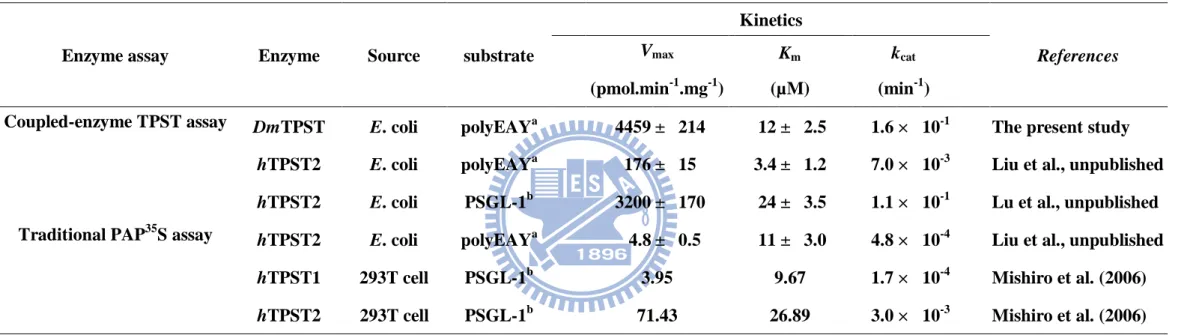
According to the previous studies, the Km values for of recombinant hTPST1
and hTPST2 obtained from cell culture (293T cells) using PSGL-1 as substrate was
9.67 and 26.9 mM, respectively; the Vmax hTPST1 and hTPST2 was 3.95 and 71.4
pmol/min/mg (the kcat was thus 1.67 × 10-4 and 2.99 × 10-3 min-1 for each other)
21
substrate polyEAY, the Km and Vmax was individually 3.4 mM and 176 pmol/min/mg;
the kcat was thus 7.0 × 10-3.(Liu et al., unpublished)(Table. 2). In this study, the Km,
Vmax, and kcat of DmTPST towards polyEAy as substrates was 11.5 mM, 4.5
nmol/min/mg, and 1.6 × 10-1, respectively (Fig. 9). Obviously, the Km values were
similar regardless of the TPST acquired from diverse sources, species and assayed by
different methods. However, the distinct Vmax measured from coupled-enzyme
reaction was higher than that of previous method for approximately 10 folds, because
the method made some modifications. Consequently the detection of polyEAY sulfate
reached to nanomolarr range (weird) and contributed to the discovery of sulfated
peptides in the future.
In the study, commercial polyEAY consisted of Glu, Ala, Tyr with random
synthesis followed the ratio of 6:3:1. Consequently, standard substrate was urgent to
be utilized in the assay. The D.melanogaster endogenous substrate, Drosulfokinin,
was selected to analyze in the DmTPST activity assay (Fig. 10). The data
demonstrated that recombinant DmTPST could not only catalyze synthetic peptide
(polyEAY) but endogenous substrate (drosulfokinin). The recombinant DmTPST will
be used further in the aspects of substrate examination, substrate screening, and
proteomic application.
22
the various purification characteristics as compared to human TPST. Furthermore, the
combination of PAPS generating system facilitated to increase the catalytic rate of
DmTPST, and define DmTPST optimal condition and kinetic parameters. This will be
beneficial to not only the aspects of fundamental researches but apply to Drosophila
23
References
1. Baeuerle, P. A., and Huttner, W. B. (1985) Tyrosine sulfation of yolk proteins 1, 2,
and 3 in Drosophila melanogaster. J. Biol. Chem. 260:6434-6439.
2. Baeuerle P. A., and Huttner, W. B. (1987) Tyrosine sulfation is a
trans-Golgi-specific protein modification. J. Cell Biol. 105: 2655–2664.
3. Baumann. (1876) Ber. Dtsch. Chem. Ges. 9:54-58.
4. Beisswanger, R., Corbeil, D., Vannier, C., Thiele, C., Dohrmann, U., Kellner, R.,
Ashman, K., Niehrs, C., and Huttner, W. B. (1998) Existence of distinct
tyrosylprotein sulfotransferase genes: molecular characterization of
tyrosylprotein sulfotransferase-2. Proc. Natl. Acad. Sci. U. S. A. 95:11134-11139.
5. Bettelheim, F. R. (1954) Tyrosine-O-sulfate in a peptide from fibrinogen. J. Am.
Chem. Soc. 76:2838–2839.
6. Chapman E., Best M.D., Hanson S.R., and Wong C.H. (2004) Sulfotransferases:
structure, mechanism, biological activity, inhibition, and synthetic utility. Angew.
Chem. Int. Ed. Engl. 43:3526-3548.
7. Colvin R.A., Campanella G.S., Manice L.A., and Luster A.D. (2006) CXCR3
requires tyrosine sulfation for ligand binding and a second extracellular loop
24
8. Dong J., Ye P., Schade A.J., Gao S., Romo G.M., Turner N.T., McIntire L.V., and López J.A. (2001) Tyrosine sulfation of glycoprotein Ibα: role of electrostatic interactions in von Willebrand factor binding. J. Biol. Chem. 276:16690-16694.
9. Farzan M., Mirzabekov T., Kolchinsky P., Wyatt R., Cayabyab M., Gerard N.P.,
Gerard C., Sodroski J., and Choe H. (1999) Tyrosine sulfation of the amino
terminus of CCR5 facilitates HIV-1 entry. Cell. 96:667–676.
10. Farzan, M., Babcock, G. J., Vasilieva, N., Wright, P. L., Kiprilov, E., Mirzabekov,
T., and Choe, H. (2002) The role of post-translational modifications of the
CXCR4 amino terminus in stromal-derived factor 1 alpha association and hiv-1
entry. J. Biol. Chem. 227:29484-29489.
11. Fong A.M., Alam S.M., Imai T., Haribabu B., and Patel D.D. (2002) CX3CR1
tyrosine sulfation enhances fractalkine-induced cell adhesion. J. Biol. Chem.
277:19418–19423.
12. Fredrickson B.J., Dong J.F., McIntire L.V., and Lo´pez J.A. (1998)
Shear-dependent rolling on von Willebrand factor of mammalian cells expressing
the platelet glycoprotein Ib-IX-V complex. Blood. 92:3684-3693.
13. Hsu, W., Rosenquist, G. L., Ansari, A. A., and Gershwin, M. E. (2005)
25
14. Huttner, W. B. (1982) Sulphation of tyrosine residues-a widespread modification
of proteins. Nature 299:273-276.
15. Jones, D. T. (2007) Improving the accuracy of transmembrane protein topology
prediction using evolutionary information. Bioinformatics 23:538-544.
16. Kehoe, J. W., and Bertozzi, C. R. (2000) Tyrosine sulfation: a modulator of
extracellular protein-protein interactions. Chem. Biol. 7:R57-61.
17. Laemmli, U. K. (1970) Cleavage of structural proteins during the assembly of
the head of bacteriophage T4. Nature 227:680-685.
18. Lee, R. W., and Huttner, W. B. (1985) (Glu62, Ala30, Tyr8)n serves as
high-affinity substrate for tyrosylprotein sulfotransferase: a Golgi enzyme. Proc.
Natl. Acad. Sci. U. S. A. 82:6143-6147.
19. Leyte A., van Schijndel H.B., Niehrs C, Huttner W.B., Verbeet M.P., Mertens K.,
and van Mourik J.A. (1991) Sulfation of Tyr1680 of human blood coagulation
factor VIII is essential for the interaction of factor VIII with von Willebrand
factor. J Biol Chem. 266:740-746.
20. Lin, E. S., and Yang, Y. S. (1998) Nucleotide binding and sulfation catalyzed by
phenol sulfotransferase. Biochem. Biophys. Res. Commun. 271:818-822.
21. Lin, W. H., and Roth, J. A. (1990) Characterization of a tyrosylprotein
26 22. Liu et al., unpublished.
23. Mann M, and Jensen O.N. (2003) Proteomic analysis of post-translational
modifications. Nat. Biotechnol. 21:255-261.
24. Marchese P., Murata M., Mazzucato M., Pradella P., De Marco L., Ware J., and
Ruggeri Z.M. (1995) Identification of three tyrosine residues of glycoprotein Ibα with distinct roles in von Willebrand factor and α-thrombin binding. J. Biol.
Chem. 270:9571- 9578.
25. Mishiro, E., Sakakibara, Y., Liu, M. C., and Suiko, M. (2006) Differential
enzymatic characteristics and tissue-specific expression of human TPST-1 and
TPST-2. J. Biochem. 140:731-737.
26. Monigatti F, Gasteiger E, Bairoch A, and Jung E. (2002) The Sulfinator:
predicting tyrosinesulfation sites in protein sequences. Bioinformatics.
18(5):769-70.
27. Monigatti, F., Hekking, B., and Steen, H. (2006) Protein sulfation analysis--A
primer. Biochim. Biophys. Acta 1764:1904-1913.
28. Moore, K. L. (2003) The biology and enzymology of protein tyrosine O-sulfation.
J. Biol. Chem. 278:24243-24246.
29. Murata M., Ware J., and Ruggeri Z.M., (1991) Site-directed mutagenesis of a soluble recombinant fragment of platelet glycoprotein Ibα demonstrating
27
negatively charged residues involved in von Willebrand factor binding. J. Biol.
Chem. 266:15474-15480.
30. Nichols R., Schneuwly S.A., Dixon J.E. (1988) Identification and
characterization of a Drosophila homologue to the vertebrate neuropeptide
cholecystokinin. J. Biol. Chem. 263:12167-12170.
31. Niehrs, C., and Huttner, W. B. (1990) Purification and characterization of
tyrosylprotein sulfotransferase. EMBO J. 9:35-42.
32. Olson P.F., Fessler L.I., Nelson R.E., Sterne R.E., Campbell A.G., Fessler J.H.
(1990) Glutactin, a novel Drosophila basement membrane-related glycoprotein
with sequence similarity to serine esterases. EMBO J. 9:1219-1227.
33. Ouyang, Y., Lane, W. S., and Moore, K. L. (1998) Tyrosylprotein
sulfotransferase: purification and molecular cloning of an enzyme that catalyzes
tyrosine O-sulfation, a common posttranslational modification of eukaryotic
proteins. Proc. Natl. Acad. Sci. U. S. A. 95:2896-2901.
34. Preobrazhensky A.A., Dragan S., Kawano T., Gavrilin M.A., G0lina I.V.,
Chakravarty L., and Kolattukudy P.E. (2000) Monocyte chemotactic protein-1
receptor CCR2B is a glycoprotein that has tyrosine sulfation in a conserved
28
35. Pouyani, T., and Seed, B. (1995) PSGL-1 recognition of P-selectin is controlled
by a tyrosine sulfation consensus at the PSGL-1 amino terminus. Cell.
83:333-343.
36. Reiter LT, Potocki L, Chien S, Gribskov M, Bier E. (2001) A systematic analysis
of human disease-associated gene sequences in Drosophila melanogaster.
Genome Res. 11(6):1114-25.
37. Sako D., Comess K.M., Barone K.M., Camphausen R.T., Cumming D.A., and
Shaw G.D. (1995) A sulfated peptide segment at the amino terminus of PSGL-1
is critical for P-selectin binding. Cell. 83, 323-331.
38. Seibert, C., and Sakmar, T. P. (2008) Toward a framework for sulfoproteomics:
Synthesis and characterization of sulfotyrosine-containing peptides. Biopolymers
90:459-477.
39. Seo J., and Lee K.J. (2004) Post-translational modifications and their biological
functions: proteomic analysis and systematic approaches. J. Biochem. Mol. Biol.
37:35-44.
40. Stone S.R., and Hofsteenge J. (1986) Kinetics of the inhibition of thrombin by
29
41. Suiko M., and Liu M.C. (1988) Change in binding affinities of 3Y1 secreted
fibronectin upon desulfation of tyrosine-O-sulfate. Biochem. Biophys. Res.
Commun. 154:1094-1098.
42. Ward C.M., Andrews R.K., Smith A.I., and Berndt M.C. (1996) Mocarhagin, a
novel cobra venom metalloproteinase, cleaves the platelet von Willebrand factor receptor glycoprotein Ibα Identification of the sulphated tyrosine/anionic
sequence Tyr-276–Glu-282 of glycoprotein Ibαas a binding site for von Willebrand factor and α-thrombin. Biochemistry. 35:4929-4938.
43. Wilkins, P.P., Moore, K.L., McEver, R.P. and Cummings, R.D. (1995) Tyrosine
sulfation of P-selectin glycoprotein ligand-1 is required for high affinity binding
to P-selectin. J. Biol. Chem. 270:22677-22680.
44. William S., Ramaprasad P., and Kasinathan C. (1997) Purification of
tyrosylprotein sulfotransferase from rat submandibular salivary glands. Arch.
Biochem. Biophys. 338:90-96.
45. Zlotnik A., Morales J. and Hedrick J.A. (1999) Recent advances in chemokines
30
Tables
Table 1. Purification of NusA-DmTPST from E. coli.
Step
Total Activity Total Protein Specific Activity Yield Purification (pmole/min) (mg) (pmole/min/mg) (%) fold
Crude extract 37325 1420 26 . 3 100 1
31
Table 2. Comparison of coupled enzyme assay-obtained kinetic characterization of DmTPST with previous radiometric assay
a
polyEAY was synthesized followed the ratio of Glu : Ala : Tyr = 6 : 3 : 1.
b
PSGL-1 was P-selectin glycoprotein ligand-1 N-terminal peptide (ATEYEYLDYDFL).
Enzyme assay Enzyme Source substrate
Kinetics References Vmax Km kcat (pmol.min-1 .mg-1) (µM) (min-1 )
Coupled-enzyme TPST assay DmTPST E. coli polyEAYa
4459 ± 214 12 ± 2.5 1.6 × 10-1 The present study
hTPST2 E. coli polyEAYa 176 ± 15 3.4 ± 1.2 7.0 × 10-3 Liu et al., unpublished
hTPST2 E. coli PSGL-1b 3200 ± 170 24 ± 3.5 1.1 × 10-1 Lu et al., unpublished Traditional PAP35S assay hTPST2 E. coli polyEAYa
4.8 ± 0.5 11 ± 3.0 4.8 × 10-4 Liu et al., unpublished
hTPST1 293T cell PSGL-1b 3.95 9.67 1.7 × 10-4 Mishiro et al. (2006)
32
Figures
Figure 1. Scheme for the determination of TPST activity. Isotope-based analysis
(35S) was used for the DmTPST assay using PAPS as the sulfuryl group donor. Biosynthesis of PAPS was catalyzed by PAPSS from ATP and SO42- as shown in Step
A. Step B showed the reaction catalyzed by TPST using tyrosylprotein as the sulfuryl group acceptor.
33
Figure 2. Bioinformatic analysis of protein sequence identity and transmembrane domain for human and Drosophila melanogaster.
The sequence pairwise alignment was performed by ClustalW (http://www.ebi.ac.uk/Tools/clustalw2/index.html) and sorted shading by BOXSHADE server (http://www.ch.embnet.org/software/BOX_form.html). The black background indicated identity to each other and the gray one meant conserved substitutions. The residue colored in red is the predicted transmembrane domain calculated by PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/psiform.html) ranged from residue 6 to 28 both for human TPST1 and TPST2, and 12 to 28 for Drosophila melanogaster TPST, respectively. The pairwise sequence identity of these TPSTs for
DmTPST/hTPST-1 (58%), DmTPST/hTPST-2 (56%), and hTPST-1/hTPST-2 (63%),
34
Figure 3. Purification of homogenous recombinant DmTPST
The protein was expressed in BL21(DE3)pLysS cells and purified through His-Tag column. Lane 1 and lane 2 was crude extract and homogenous DmTPST, respecticely, and lane 3 was standard protein marker.
35
Figure 4. The protein of LC-MS-MS fingerprinting analysis was identified as
DmTPST.
The excised spot from SDS-PAGE was identified as DmTPST by LC-MS-MS. The sequence (red) obtained from mass fingerprinting was mapped to the protein sequence with high confidence. The result was particularly described in Appendix 8 and
36
Figure 5. Autoradiography of [35S]-labeled polyEAY catalyzed by DmTPST.
Lanes 1 to 3 were controlled reactions by the presence of DmTPST w/o PAPSS (lane 1), polyEAY (lane 2) and DmTPST (lane 3), respectively. Lane 4 using polyEAY as substrate proceeding sulfation reaction. The arrowheads indicated the [35S]sulfated polyEAY peptides and [35S]sulfate.
37
Figure 6. Effective range of DmTPST assay.
polyEAY sulfation catalyzed by the variable amount of recombinant DmTPST (from 0.5 to 20 g) was determined under the standard condition. Each point and bar represented the mean and SD, respectively, obtained from three experiments.
DmTPST amount (g) 0 5 10 15 20 25 polyE AY sulfat e ( nmol/m in) 0 5 10 15 20 25 30 DmTPST amount(g) 0 1 2 3 4 5 polyE AY sulfat e( nmol/m in) 0 2 4 6 8 10 12 14 16
38
Figure 7. Time course of the activity of recombinant DmTPST.
Time course effected on the activity of DmTPST. Activities of DmTPST were measured in different time (15, 30, 45, 60, 90, 120 min) under the standard condition. Each point and bar represented of three experiments.
Time(min)
0 20 40 60 80 100 120 140poly
E
A
Y
su
lfate(nm
ol/
m
g)
60 80 100 120 140 160 180 200 22039
Figure 8. pH profile effected on the activity of recombinant DmTPST.
The pH profile of DmTPST activity. TPST activities were measured in 50 mM buffer at selected pHs (MES for pH 5.5, 6.0, 6.5, potassium phosphate for pH 6.5, 7.0, 7.5, and Tris for pH 7.5, 8.0, and 8.5) under the standard condition. Each point and bar represented the mean SD of three experiments.
pH
5.0 5.5 6.0 6.5 7.0 7.5 8.0 8.5 9.0Specific Acti
vity
(nmole/min/m
g)
0.0 0.5 1.0 1.5 2.0 2.5 3.0 3.5 4.0 MES Potassium phosphate Tris40
Figure 9. Kinetics of DmTPST using polyEAY as substrate.
The DmTPST kinetics activities were determined under the standard condition with various polyEAY concentration from 0.3125 to 160 M. Each point and bar represented the mean SD of three experiments. The data indicated the Km and Vmax
was 11.5 ± 2.5 mM and 4.5 ± 0.2 nmole/min/mg, respectively.
Michaelis-Menten
[polyEAY] (
M)
0 20 40 60 80 100 120 140 160 180Specific Acti
vity
(nmol/m
in/mg)
0 1 2 3 4 5 641
Figure 10. Autoradiography of [35S]-labeled Drosulfakinin catalyzed by
DmTPST.
Lanes 1 to 4 were controlled reactions in the absence of PAPSS (lane 1), substrate (lane 2), and DmTPST (lane 3), respectively. Lane 4 using polyEAY as substrate proceed sulfation reaction was positive control, and lane 5 was full reaction that using drosulfakinin as substrate. The arrowhead indicated the [35S]sulfated drosulfakinin peptides and [35S]sulfate.
42
Appendix
Appendix 1. Schematic representation of protein modifications related to the regulation of biological processes. (Adapted from Seo et al. 2004)
43
Appendix 2. Some common and important post-translation modifications (Mann
et al., 2003).
44
Appendix 3. General ST-catalyzed reaction with PAPS as the cosubstrate
45
Appendix 4. Sulfate activation and tyrosine O-sulfation. Inorganic sulfate enters
the cell by the action of one of several sulfate transporters. Once in the cytosol, sulfate is then activated by the action of one of two PAPS synthases (PAPSS1 or PAPSS2). These bifunctional enzymes contain a C-terminal ATP sulfurylase domain and an N-terminal adenosine phosphosulfate (APS) kinase domain. In the first step of sulfate activation, ATP and inorganic sulfate are converted to APS and pyrophosphate by ATP sulfurylase. APS is then channeled directly between the ATP sulfurylase and APS kinase active sites. In the second step catalyzed by the APS kinase domain, a second ATP is consumed to phosphorylate the 3’-hydroxyl of the ribose ring of APS to yield PAPS and ADP. PAPS is then transported into the Golgi lumen by a PAPS translocase that has been purified but not yet cloned. This transporter functions via an antiporter mechanism with PAP as the returning ligand. Once inside the Golgi lumen PAPS acts as the sulfate donor for TPSTs and all other carbohydrate sulfotransferases, and the sulfated products are either secreted or retained in the membrane of lysosomes, secretory vesicles, and/or the plasma membrane. TGN, trans-Golgi network. (Moore, 2003).
46
Appendix 5. Schematic representation of cell entry by HIV-1 following
47
Appendix 6. Tyrosine sulfation plays an important role in the immune response.
(a) Leukocytes roll upon, adhere to and transmigrate between endothelial cells at sites of inflammation. P-selectin and its ligand, PSGL-1, are often required for this process. (b) PSGL-1 is a mucinlike glycoprotein that appears to be an extended rod shape in
vivo. The extreme amino terminus of PSGL-1 carries three tyrosine sulfation sites,
shown in yellow. These sulfate esters, and specific glycans on PSGL-1, are key binding determinants for P-selectin (Kehoe and Bertozzi, 2000).
48
49
Mascot Search Results
Protein View
Match to: TPST_DROME Score: 112
Protein-tyrosine sulfotransferase OS=Drosophila melanogaster GN=Tango13 PE=2 SV=2
Found in search of MI806075.pkl
Nominal mass (Mr): 58013; Calculated pI value: 7.66
NCBI BLAST search of TPST_DROME against nr
Unformatted sequence string for pasting into other applications Taxonomy: Drosophila melanogaster
Cleavage by Trypsin: cuts C-term side of KR unless next residue is P Sequence Coverage: 4%
Matched peptides shown in Bold Red
1 MRLPYRNKKV TLWVLFGIIV ITMFLFKFTE LRPTCLFKVD AANELSSQMV 51 RVEKYLTDDN QRVYSYNREM PLIFIGGVPR SGTTLMRAML DAHPDVRCGQ 101 ETRVIPRILQ LRSHWLKSEK ESLRLQEAGI TKEVMNSAIA QFCLEIIAKH 151 GEPAPRLCNK DPLTLKMGSY VIELFPNAKF LFMVRDGRAT VHSIISRKVT 201 ITGFDLSSYR QCMQKWNHAI EVMHEQCRDI GKDRCMMVYY EQLVLHPEEW 251 MRKILKFLDV PWNDAVLHHE EFINKPNGVP LSKVERSSDQ VIKPVNLEAM 301 SKWVGQIPGD VVRDMADIAP MLSVLGYDPY ANPPDYGKPD AWVQDNTSKL 351 KANRMLWESK AKQVLQMSSS EDDNTNTIIN NSNNKDNNNN QYTINKIIPE 401 QHSRQRQHVQ QQHLQQQQQQ HLQQQQHQRQ QQQQQREEES ESEREAEPDR 451 EQQLLHQKPK DVITIKQLPL AGSNNNNINN NINNNNNNNN IMEDPMADT
Appendix 8. MS analysis of DmTPST
../data/20100630/Ft TPST_DROME 1 false 44
0 0.05 0 1 1025
50
Mascot Search Results
Protein View
Match to: NUSA_ECOLI Score: 321
Transcription elongation protein nusA OS=Escherichia coli (strain K12) GN=nusA PE=1 SV=1
Found in search of MI806075.pkl
Nominal mass (Mr): 54837; Calculated pI value: 4.53
NCBI BLAST search of NUSA_ECOLI against nr
Unformatted sequence string for pasting into other applications Taxonomy: Escherichia coli K-12
Cleavage by Trypsin: cuts C-term side of KR unless next residue is P Sequence Coverage: 10%
Matched peptides shown in Bold Red
1 MNKEILAVVE AVSNEKALPR EKIFEALESA LATATKKKYE QEIDVRVQID 51 RKSGDFDTFR RWLVVDEVTQ PTKEITLEAA RYEDESLNLG DYVEDQIESV
101 TFDRITTQTA KQVIVQKVRE AERAMVVDQF REHEGEIITG VVKKVNRDNI 151 SLDLGNNAEA VILREDMLPR ENFRPGDRVR GVLYSVRPEA RGAQLFVTRS
201 KPEMLIELFR IEVPEIGEEV IEIKAAARDP GSRAKIAVKT NDKRIDPVGA 251 CVGMRGARVQ AVSTELGGER IDIVLWDDNP AQFVINAMAP ADVASIVVDE 301 DKHTMDIAVE AGNLAQAIGR NGQNVRLASQ LSGWELNVMT VDDLQAKHQA 351 EAHAAIDTFT KYLDIDEDFA TVLVEEGFST LEELAYVPMK ELLEIEGLDE
401 PTVEALRERA KNALATIAQA QEESLGDNKP ADDLLNLEGV DRDLAFKLAA 451 RGVCTLEDLA EQGIDDLADI EGLTDEKAGA LIMAARNICW FGDEA
Appendix 9. MS analysis of NusA
../data/20100630/Ft NUSA_ECOLI 1 false 44
0 0.05 0 1 1025



![Figure 5. Autoradiography of [ 35 S]-labeled polyEAY catalyzed by DmTPST.](https://thumb-ap.123doks.com/thumbv2/9libinfo/8763208.208684/49.892.275.628.165.457/figure-autoradiography-s-labeled-polyeay-catalyzed-dmtpst.webp)