行政院國家科學委員會專題研究計畫 期中進度報告
全基因體化分析克雷白氏肺炎桿菌纖毛基因組的表現(1/3)
計畫類別: 個別型計畫 計畫編號: NSC94-3112-B-009-004- 執行期間: 94 年 05 月 01 日至 95 年 04 月 30 日 執行單位: 國立交通大學生物科技學系(所) 計畫主持人: 彭慧玲 計畫參與人員: 黃盈蓉,吳健誠,廖心瑋,黃登魁,廖朝陽 報告類型: 完整報告 報告附件: 出席國際會議研究心得報告及發表論文 處理方式: 本計畫可公開查詢中 華 民 國 95 年 5 月 9 日
行政院國家科學委員會補助專題研究計畫
■ 成 果 報 告
□期中進度報告
Genome wide analysis of expression of the fimbrial operons in
Klebsiella pneumoniae
計畫類別:■ 個別型計畫 □ 整合型計畫
計畫編號:NSC 94-3112-B-009-004-
執行期間: 九十五 年 五 月 一 日 至 九十六 年 四 月 三十 日
計畫主持人:彭慧玲
共同主持人:
計畫參與人員:黃盈蓉,吳健誠,廖心瑋,黃登魁,廖朝陽
成果報告類型(依經費核定清單規定繳交):□精簡報告 ■完整報告
本成果報告包括以下應繳交之附件:
□赴國外出差或研習心得報告一份
□赴大陸地區出差或研習心得報告一份
■出席國際學術會議心得報告及發表之論文各一份
□國際合作研究計畫國外研究報告書一份
處理方式:除產學合作研究計畫、提升產業技術及人才培育研究計畫、
列管計畫及下列情形者外,得立即公開查詢
□涉及專利或其他智慧財產權,■一年□二年後可公開查詢
執行單位:
中 華 民 國 九十五 年 四 月 三十 日
TABLE OF CONTENTS
I. Abstracts
Ia. Abstract in Chinese
page IIIb. Abstract in English
page IIIII. Background and Significance
page 1III. Material and Methods
page 3IV. Progress Report and Discussion
page 6V. Literature Cited
page 18VI. Self-Assessment
page 20Appendix I.
Report for attending the 158th society for general Microbiology
(SGM) in Europe (held in University of Warwick, Coventry, UK)
April 3-6, 2006
Appendix II.
Poster for attending the 158th society for general Microbiology
(SGM) in Europe (held in University of Warwick, Coventry, UK)
April 3-6, 2006
I. Abstract
Ia. Abstract in Chinese
關鍵詞:克雷白氏肺炎桿菌,纖毛,黏附因子,黏附特異性 在克雷白氏肺炎桿菌NTUH K-2044 基因體中,除了已知的第一型及 第三型纖毛外,我們另外發現了七套未被發表過的纖毛基因組,分別命名 為 kpa,kpb,kpc,kpd,kpe,kpf 和 kpg。為了了解這九套纖毛基因組在 黏附特異性及表現調控上的異同,我們在過去一年完成了下列工作: 1. 以 ClustalW 工具分別進行纖毛蛋白質的序列分析,可發現這九套纖毛之 間並沒有明顯的親緣關係。 2. 纖毛基因存在分佈率的研究結果顯示,不同來源的123株克雷白氏肺炎桿 菌分離株,大多都含有kpa、kpe、kpg、fim, 以及mrk纖毛基因組 (87% to 100%)。此外,在35株K1莢膜血清型分離株中,有32株具有相同的纖毛基 因組合。 然而,這九套纖毛基因組的存在率與疾病種類並沒有明顯的關 聯性。 3. 為了瞭解克雷白氏肺炎桿菌調控如何這九種纖毛的表現,我們已分別將這 九套纖毛基因子的起使子轉殖到LacZ通報系統,以及製備了九種Balb/c小 鼠抗體,可分別偵測九種纖毛的主要結構蛋白質。 4. (1) 九套纖毛基因組已分別自克雷白氏肺炎桿菌 NTUH K-2044中選殖出 來,並且以不會生成纖毛的大腸桿菌為宿主,分別表現於大腸桿菌表面。 (2) 我們證明第一型及第三型纖毛的表現有互相拮抗的效應 (3) MrkD黏 附因子基因序列差異會影響第三型纖毛的生成和特異性表現 (4) 我們證 明了MrkF為第三型纖毛的結構蛋白質之一。
I. Abstract
Ib. Abstract in English
Keywords:Klebsiella pneumoniae, fimbriae, ahesins, adherence specificities
In the genome of Klebsiella pneumoniae NTUH K-2044, in addition to the identified type 1 and type 3 fimbriae, the seven novel fimbrial gene clusters were named kpa, kpb, kpc, kpd, kpe, kpf, and kpg. To understand whether these fimbrial operons function synergistically or are regulated differentially, we have accomplished several tasks in the past year as followings:
1. Multiple sequence alignment of the fimbrial genes by ClustalW revealed that the four major components of the nine fimbrial operons are distinct to each other.
2. The prevalence analysis of the nine fimbrial gene clusters among 123 K.
pneumoniae clinical isolates from various types of disease indicated that kpa, kpe, kpg, fim, and mrk genes were contained in most of the isolates (87% to
100%). In addition, 32 of 35 K1 isolates appeared to possess identical repertoire of fimbrial operons. However, no obvious correlation between the fimbrial types and diseases could be identified.
3. To characterize each of the fimbriae and investigate if these fimbriae express differentially in the bacteria, each of the putative promoters has been cloned respectively into a LacZ reporter vector and polyclonal antibodies targeting to the major subunit of each of the fimbriae have also been raised from Balb/c mice.
4. (1) Each of the fimbrial operons have been isolated by PCR-based cloning from
K. pneumoniae NTUH K-2044 and the fimbriae displayed on the surface of an
afimbriated E. coli. (2) A cross-talk regulation between type1 and type 3 fimbriae has been demonstrated. (3) Allelic variation of mrkD adhesin affected fimbriation and activity of type 3 fimbriae (4) MrkF was demonstrated to be a component of type 3 fimbriae.
II. Background and Significance
The biosynthesis and binding properties of type 1 fimbriae have been well studied in
Escherichia coli (12, 13, 14, 15). Although with less information, type 1 fimbriae
have also been isolated and characterized in K. pneumoniae (4, 7, 8). Type 3 fimbrial adhesin, referred to as mannose-resistant Klebsiella-like (MR/K) hemagglutinin, has been shown to be produced by a few of K. pneumoniae strains (1, 8, 9). In addition to the two fimbrial adhesins, three more including the fimbrial antigen KPF-28, the afimbrial adhesin CF29K, and an afimbrial adhesin composed of capsule-like extracellular material have recently been reported in Klebsiella isolates (2, 3, 6).
Adhesin specificity plays a major role in determining the host range and tissue tropism in bacterial infection (17). Depending on the expression of type 1 or P fimbriae, UPEC (uropathogenic E. coli) vary significantly in their abilities to colonize and persist within the bladder or kidneys (22, 23). PapB, a regulator for phase variation of P fimbriae, and its functional homologue, SfaB of S fimbriae, both were shown to be able to increase the frequency of ON-to-OFF phase for type 1 fimbriae (10). This indicated the presence of a regulatory network in controlling differential expression of type 1-, P-, and S-fimbriae in UPEC. In E. coli, type 1 fimbriae has been demonstrated to be essential for the bacteria in establishment and persistence of urinary tract infections (12, 22). Most likely, the fimbriae are also required for K. pneumoniae to colonize urinary tract. In addition to uroepithelial cells, the adherence to respiratory epithelia, intestinal and endothelial cells of the K.
pneumoniae strains expressing type 3 fimbriae have been demonstrated (16).
However, the specificity and regulation of fimbrial adherence in K. pneumoniae are largely unknown.
Prevalence analysis of Salmonella enterica sp. Typhi isolates revealed a unique repertoire of fimbrial gene clusters possibly due to specific selective pressures (20). Expression analysis using flow cytometry of eleven S. Typhimurium fimbrial operons indicated further that in vivo growth conditions drastically alter the
expression of repertoire of fimbrial antigens (11). Thus, differential expression of the nine fimbriae for K. pneumoniae to adhere to different cells or abiotic surfaces, or for the bacterial self-aggregation or biofilm formation could be foreseen. Using bioinformatic analysis, nine sets of fimbriae encoding operons of the chaperone-usher assembly pathway which includes type 1-, type 3- and seven novel-type fimbriae, namely kpa, kpb, kpc, kpd, kpe, kpf, and kpg in the genome of
K. pneumoniae NTUH K-2044 (http://genome.nhri.org.tw/kp/index.php), a K1
isolate of pyogenic liver abscess (5), were identified. The specific aims proposed to be accomplished included:
(1) Analysis of the fimbrial gene clusters in K. pneumoniae using the tools of bioinformatics
(2) Investigation of the prevalence of the fimbrial gene clusters in K.
pneumoniae clinical isolates
(3) Determination of the regulatory control of expression of the fimbrial operons
(4) Characterization of the adherence specificities displayed by each of the fimbrial adhesins
III. Material and Methods
Bacterial strains and growth condition. K. pneumoniae NTUH-K2044, of K1
serotype, was provided by Dr. Jin-Town Wang from National Taiwan University Hospital (5). This is a highly invasive and hypermucous strain orignally isolated from
the blood of a 40 year old male patient at suffering from community-acquired primary liver abscess and metastatic meningitis (5). The K. pneumoniae NTUH-K2044
genome sequence of about 5.5 Mbp was determined by Dr. Shih-Feng Tsai’s group in Division of Molecular and Genomic Medicine, National Health Research Institutes
(manuscript in submission, the sequences will be released after the manuscript accepted for publication). Clinical isolates of K. pneumoniae used in this study were
recovered from different tissue specimens of patients with a variety of infections at the Veteran General Hospital, Taipei, from 1991 to 1998. The strains were identified
and serotype determined. The bacteria were grown in Luria-Bertani (LB) broth or agar at 37°C and stored at -80°C before use.
PCR amplification for prevalence analysis. The PCR mixture contained 20 mM
Tris-HCl (pH 8.4), 50 mM KCl, 1.5 mM MgCl2, 200 μM (each) deoxynucleoside
triphosphates (dATP, dCTP, dGTP, and dTTP), and 1 U of recombinant Taq DNA
polymerase (Violet Bioscience Inc.) along with each of the K. pneumoniae genomic DNA and the specific primers. The primer pairs designed for the respective adhesin
and pilin encoding genes of each fimbrial operons are shown in Table 1. The amplification cycle consisted of an initial 1 min hold at 95°C followed by 35 cycles
of 95°C for 1 min, 50°C for 1 min, and 72°C for 1 min, and finally an elongation step for 10 min at 72°C. The amplified PCR product was then analyzed by
Antiserum preparation. The coding regions corresponding to FimA and MrkA, the
major pilin subunit of type 1 fimbriae and type 3 fimbriae, were amplified by PCR
and cloned into expression vector pET30, respectively. The expression plasmid was then transformed into E. coli Nova-Blue (DE3) and overexpression of the
recombinant pilin was induced by addition of 0.5 mM IPTG (isopropyl-β-D-thiogalactopyranoside). The MrkA and FimA proteins fusion with
His-tag were then purified using a nickel charged resin (Novagen, Madison, WI). In order to raise antibody, five-week-old female BALB/c mice, purchased from the
animal center of National Taiwan University, were immunized intraperitoneally with 5 μg of the purified MrkA or FimA. Ten days later, the mice were immunized again
with 5 μg of the MrkA or FimA protein. Finally, the antisera were obtained by intracardic puncture.
Western blot analysis of the expression of type 1 and type 3 fimbriae. The
clinical isolates of K. pneumoniae were grown overnight in either LB, for optimal
expression of type 1 fimbriae, or GCAA (minimal medium supplemented with 1% glycerol and 0.3% casamino acids) broth for optimal expression of type 3 fimbriae.
Total cellular proteins of the bacteria were resolved by SDS-PAGE and electrophoretically transferred from the gels onto PVDF membranes
(ImmobilonTM-P, Millipore). The membranes were then blocked with 5% skimmilk at room temperature for 1 h, and washed 3 times with 1 X phosphate-buffered saline
(PBS). Subsequently, the membranes were incubated with diluted anti-FimA or anti-MrkA serum at room temperature for 1 h. After 3 washes with 1 X PBS, a
3000-fold diluted alkaline phosphatase-conjugated anti-mouse immunoglobulin G was added and the incubation continued for one more hour. The blot was again
BCIP (5-bromo-4-chloro-3-indolyl phosphate) and NBT (Nitro blue tetrazolium).
Statistical methods. Fisher's exact testwas used to determine the statistical
IV. Progress Report and Discussion
Specific aim 1- Analysis of the fimbrial gene clusters in K. pneumoniae using the tools of bioinformatics. Using HMMER to search for the Pfam fimbrial
family and BLAST analysis of the K. pneumoniae NTUH K-2044 genome, nine
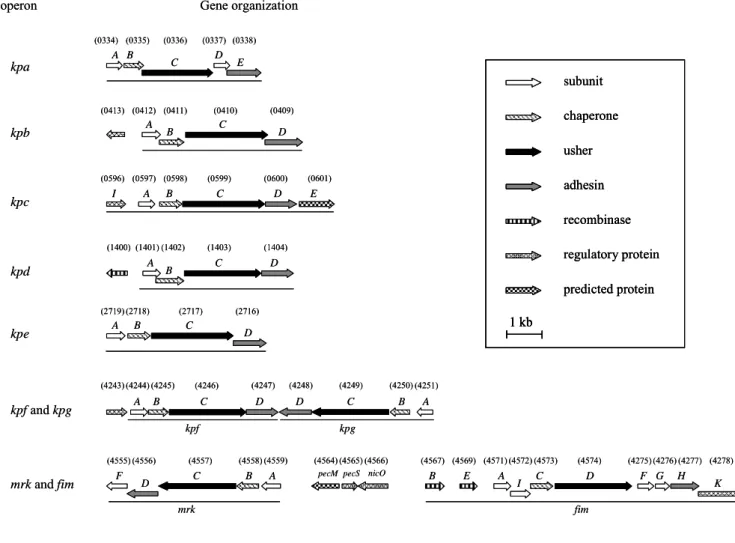
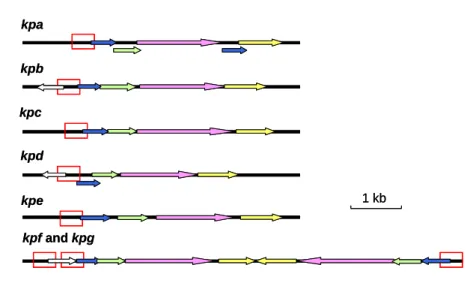
distinct putative fimbrial operons including type 1 and type 3, and seven novel types of fimbriae, namely kpa, kpb, kpc, kpd, kpe, kpf, and kpg were identified (Fig. 1).
Each of the fimbrial operons contains the four essential genes for fimbriae biosynthesis, which are pilin, adhesion, chaperone, and usher encoding genes.
Multiple sequence alignment of the fimbrial operons by ClustalW indicated that they share amino acid sequence positivities of 26.5% and 36.4%, respectively, for the
genes encoding adhesin and pilin; 49.3% and 55.4% for those encoding usher and chaperone.
It is of interest to note that the fim and mrk gene clusters, encoding respectively type 1 and 3 fimbriae, are physically linked in the K. pneumoniae NTUH-K2044
genome. In between the two operons, two genes encoding putative regulators with amino acid sequence identities of 61% and 65% respectively to Erwinia
chrysanthemi PecS and PecM, the regulators of virulence control in the bacteria
were identified. Since the gene organization was also conserved in the genome of K.
pneumoniae MGH78578 (http://genome.wustl.edu/tools/blast/), the possibility of
cross-talk regulation and regulatory roles of the PecS and PecM homologs for the
expression of the two fimbriae has also been investigated (described in the section of specific aim 4).
operon Gene organization subunit chaperone usher adhesin recombinase regulatory protein predicted protein 1 kb kpa A B C D E kpb A B C D kpd A B C D kpc I A B C D E A B C kpe D kpf and kpg kpf kpg A B C D D C B A mrk and fim mrk fim F C B A D B E A I C D F G H K pecM pecS 334) (0335) (0336) (0337) (0338) (0413) (0412) (0411) (0410) (0409) 596) (0597) (0598) (0599) (0600) (0601) (1400) (1401) (1402) (1403) (1404) 719) (2718) (2717) (2716) 243) (4244) (4245) (4246) (4247) (4248) (4249) (4250) (4251) (4555) (4556) (4557) (4558) (4559) (4564) (4565)(4566) (4567) (4569) (4571)(4572) (4573) (4574) (4275) (4276) (4277) (4278) nicO
operon Gene organization
(0 (0 (2 (4 subunit chaperone usher adhesin recombinase regulatory protein predicted protein 1 kb subunit chaperone usher adhesin recombinase regulatory protein predicted protein 1 kb 1 kb kpa A B C D E kpb A B C D kpd A B C D kpc I A B C D E A B C kpe D kpf and kpg kpf kpg A B C D D C B A mrk and fim mrk fim F C B A D B E A I C D F G H K pecM pecS 334) (0335) (0336) (0337) (0338) (0413) (0412) (0411) (0410) (0409) 596) (0597) (0598) (0599) (0600) (0601) (1400) (1401) (1402) (1403) (1404) 719) (2718) (2717) (2716) 243) (4244) (4245) (4246) (4247) (4248) (4249) (4250) (4251) (4555) (4556) (4557) (4558) (4559) (4564) (4565)(4566) (4567) (4569) (4571)(4572) (4573) (4574) (4275) (4276) (4277) (4278) nicO (0 (0 (2 (4
FIG. 1. Fimbrial gene clusters of the chaperone-usher-dependent-dependent
assembly class in K. pneumoniae NTUH-K2044. The designation of genes and locus
tag (KP number) of open reading frames annotated in the K. pneumoniae NTUH-K2044 genome were indicated. A total number of nine fimbrial gene clusters
and the adjacent regulatory proteins are as shown, and each of the putative fimbrial operons is underlined.
The BLAST analysis also revealed that, except the kpbABCD and kpeABCD operons, homologs could be identified in other bacteria for kpa, kpc, kpd, kpg, and
kpf gene clusters. The kpaABCDE operon, is likely an orthologue of Salmonella
Typhimurium LT2 sthABCDE operon, which has been reported to be required for
intestinal persistence in mice. In kpa operon,both kpaA and kpaD genes appeared to be capable of encoding a fimbrial subunit suggesting that they are respectively the
major and minor subunits of the fimbrial rod. The kpcABCD are homologous to the putative fimbrial genes, plu2159 to plu2156, of Photorhabdus luminescens subsp.
laumondii TTO1. The kpdABCDencodes a homologous operon of Reut02000958 to Reut02000961 in Ralstonia metallidurans CH34 (unfinished sequence, GenBank:
AAAI00000000). The physically linked kpfABCD operon and kpgABCD genes appeared to be respectively an orthologue of Edwardsiella tarda KG8401 etfABCD
operon, and an orthologue of the fimbrial operon, plu2159 to plu2156, in P.
luminescens subsp. laumondii TTO1.
Upstream of the kpbABCD operon, a divergently transcribed gene encoding a
transcriptional regulator with a DNA-binding domain was identified and designated
kpbR (Fig. 1). A DNA recombinase encoding gene, namely kpcI, located upstream of kpcABCD, was found to be an orthologue of several fimbrial invertases including
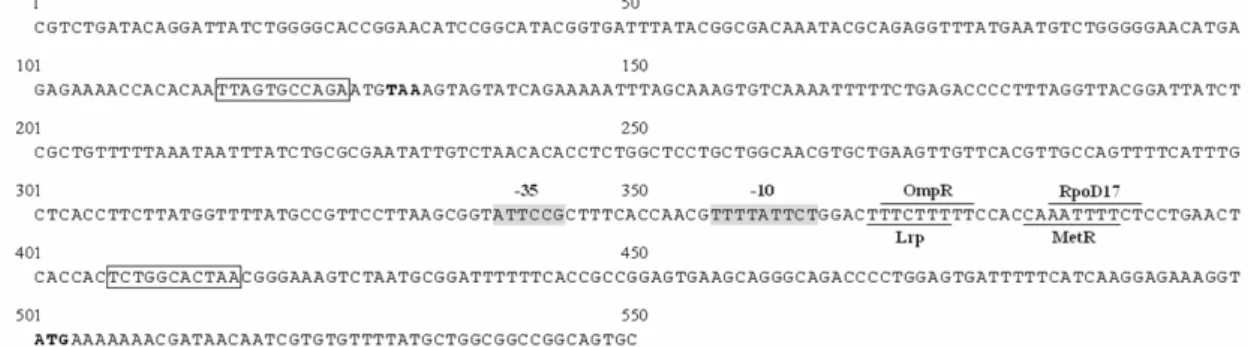
MrpI, MrxI (AAM91928) and MrfI. As shown in Fig. 2, an 11-bp inverted repeat
was identified in the putative promoter region of kpc operon, impling a phase-variation control by DNA inversion for the operonexpression. Moreover, TCS
(two component system) response regulator encoding genes, namely kpdR and kpfR, were found to be located upstream of kpdABCD and kpfABCD operons (Fig. 1).
FIG. 2. The DNA sequence of the putative promoter region of kpc operon. 500 bp
upstream and 50 bp downstream of the kpcA translation start codon ATG (bold type)
are as shown. The 11 bp inverted repeats are indicated as boxes. The predicted -10 and -35 promoter regions are shadowed. Four predicted transcription factor binding
sites are each overlined or underlined. The kpcI translation stop codon TAA is also shown as bold type.
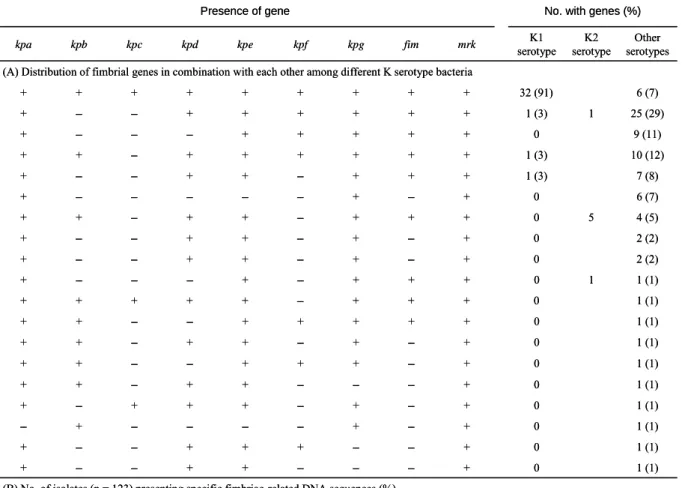
Specific aim 2- Investigation of the prevalence of the fimbrial gene clusters in K. pneumoniae clinical isolates. Prevalence analysis via PCR detection of the
respective pilin and adhesin encoding genes in 123 K. pneumoniae clinical isolates
from various types of diseases (collected by Dr. CP Fung from the Veteran General Hospital Taipei) revealed that kpa, kpe, kpg, fim, and mrk genes were contained in
most of the isolates (87% to 100%). As shown in Table 1, the distribution of kpb, kpc,
kpd, and kpf genes were 50%, 32%, 83%, and 70%, respectively. In addition, 32 of
35 K1 isolates appeared to possess identical repertoire of fimbrial operons. No obvious correlation between the fimbrial types and diseases could be identified.
(B) No. of isolates (n = 123) presenting specific fimbriae-related DNA sequences (%)
(A) Distribution of fimbrial genes in combination with each other among different K serotype bacteria
No. with genes (%) Presence of gene 122 (99) + + – + + + + + + + + + + + + + + + + kpa 6 (7) 32 (91) + + + + + + + + 123 (100) 106 (86) 120 (98) 88 (72) 116 (94) 103 (84) 40 (33) 64 (52) 2 (2) 0 + – + – + + – – 1 (1) 0 + + + – + + + + 1 (1) 1 (1) 1 (1) 1 (1) 1 (1) 1 (1) 1 (1) 1 (1) 1 (1) 2 (2) 4 (5) 6 (7) 7 (8) 10 (12) 9 (11) 25 (29) Other serotypes 0 + – + – + + – – 5 0 + + + – + + – + 0 + – + – – – – – 1 (3) + + + – + + – – 1 (3) + + + + + + – + 0 + + + + + – – – 1 1 (3) + + + + + + – – 0 + – – – + + – – 0 + – – + + + – – 0 + – + – – – – + 0 + – + – + + + – 0 + – – – + + – + 0 + – + + + – – + 0 + – + – + + – + 0 + + + + + – – + 1 0 + + + – + – – – K2 serotype K1 serotype mrk fim kpg kpf kpe kpd kpc kpb
(B) No. of isolates (n = 123) presenting specific fimbriae-related DNA sequences (%)
(A) Distribution of fimbrial genes in combination with each other among different K serotype bacteria
No. with genes (%) Presence of gene 122 (99) + + – + + + + + + + + + + + + + + + + kpa 6 (7) 32 (91) + + + + + + + + 123 (100) 106 (86) 120 (98) 88 (72) 116 (94) 103 (84) 40 (33) 64 (52) 2 (2) 0 + – + – + + – – 1 (1) 0 + + + – + + + + 1 (1) 1 (1) 1 (1) 1 (1) 1 (1) 1 (1) 1 (1) 1 (1) 1 (1) 2 (2) 4 (5) 6 (7) 7 (8) 10 (12) 9 (11) 25 (29) Other serotypes 0 + – + – + + – – 5 0 + + + – + + – + 0 + – + – – – – – 1 (3) + + + – + + – – 1 (3) + + + + + + – + 0 + + + + + – – – 1 1 (3) + + + + + + – – 0 + – – – + + – – 0 + – – + + + – – 0 + – + – – – – + 0 + – + – + + + – 0 + – – – + + – + 0 + – + + + – – + 0 + – + – + + – + 0 + + + + + – – + 1 0 + + + – + – – – K2 serotype K1 serotype mrk fim kpg kpf kpe kpd kpc kpb
TABLE 1. Frequency and repertoire of fimbrial genes among K. pneumoniae
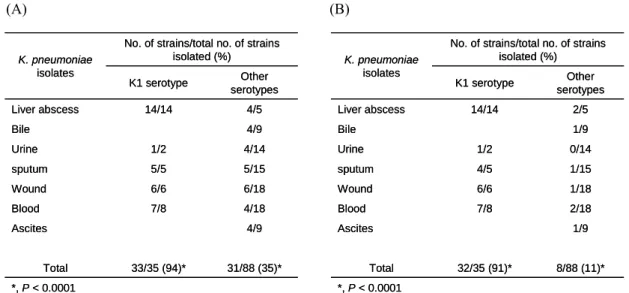
Nevertheless, kpb and kpc gene clusters were found to be more prevalent (P < 0.0001) in the isolates of K1 serotype (Table 2).
(A) (B) *, P < 0.0001 31/88 (35)* 33/35 (94)* Total Other serotypes K1 serotype 4/9 Ascites 4/18 7/8 Blood 6/18 6/6 Wound 5/15 5/5 sputum 4/14 1/2 Urine 4/9 Bile 4/5 14/14 Liver abscess
No. of strains/total no. of strains isolated (%) K. pneumoniae isolates *, P < 0.0001 31/88 (35)* 33/35 (94)* Total Other serotypes K1 serotype 4/9 Ascites 4/18 7/8 Blood 6/18 6/6 Wound 5/15 5/5 sputum 4/14 1/2 Urine 4/9 Bile 4/5 14/14 Liver abscess
No. of strains/total no. of strains isolated (%) K. pneumoniae isolates *, P < 0.0001 8/88 (11)* 32/35 (91)* Total Other serotypes K1 serotype 1/9 Ascites 2/18 7/8 Blood 1/18 6/6 Wound 1/15 4/5 sputum 0/14 1/2 Urine 1/9 Bile 2/5 14/14 Liver abscess
No. of strains/total no. of strains isolated (%) K. pneumoniae isolates *, P < 0.0001 8/88 (11)* 32/35 (91)* Total Other serotypes K1 serotype 1/9 Ascites 2/18 7/8 Blood 1/18 6/6 Wound 1/15 4/5 sputum 0/14 1/2 Urine 1/9 Bile 2/5 14/14 Liver abscess
No. of strains/total no. of strains isolated (%)
K. pneumoniae
isolates
TABLE 2. Prevalence and distribution of kpb (A) and kpc (B) genes in various
isolates of K. pneumoniae.
The results indicated that the specific combination and regulation of these fimbrial
operons may play important roles in K. pneumoniae K1 pathogenesis. Interestingly, western blotting hybridization using anti-FimA and anti-MrkA antibodies,
respectively, revealed that only 15.7% and 46.3% of the isolates expressed in vitro the major subunit of type 1 and type 3 fimbriae. Nevertheless, type 1 fimbriae were
found to be prevalently expressed (71%) in liver abscess isolates of K1 serotype suggesting a not yet identified role of type 1 fimbrial expression for the infection of
62 (50) 43 (3)
10 (2) 14 (2)
Type 3 fimbriae
% of isolates expressing fimbriae under the specific conditions (No.)
1 (8) 14 (1) 0 (0) 71 (10) Type 1 fimbriae Other serotypes (81) K2 isolates (7) K1 isolates, nonKLA (21) K1 isolates, KLA (14)
K. pneumoniae isolates (No.)
Expression of fimbriae 62 (50) 43 (3) 10 (2) 14 (2) Type 3 fimbriae
% of isolates expressing fimbriae under the specific conditions (No.)
1 (8) 14 (1) 0 (0) 71 (10) Type 1 fimbriae Other serotypes (81) K2 isolates (7) K1 isolates, nonKLA (21) K1 isolates, KLA (14)
K. pneumoniae isolates (No.)
Expression of fimbriae
TABLE 3. In vitro expression of type 1 and type 3 fimbriae
Specific aim 3- Determination of the regulatory control of expression of the fimbrial operons. (1) In order to measure the expression of each of the fimbriae, the
putative promoter regions of each of the nine fimbrial gene clusters have been
isolated from K. pneumoniae NTUH-K2044, and cloned into the reporter plasmid placZ15 containing a promoterless lacZ gene, which allow determine the signals that
affect the fimbriae expression and the promoter activity of each of the fimbriae (Fig. 3). kpa kpb kpc kpd kpe kpf and kpg 1 kb kpa kpb kpc kpd kpe kpf and kpg 1 kb
FIG. 3. Schematic diagram of the putative promoters of kpa, kpb, kpc, kpd, kpe, kpf,
The activity of the promoters including P-kpa, P- kpb, P- kpd, P- kpf, and P- kpg are
being analyzed in the presence of different cultural conditions such as changes of
nutrient sources, growth temperature, osmolarity, and iron concentration. (2) The heterologous expression constructs in Escherichia coli for each of the pilins have
been generated, the recombinant proteins purified, and several specific polyclonal antibodies obtained.
Specific aims 4- Characterization of the adherence specificities displayed by each of the fimbrial adhesins. (1) To characterize each of the fimbriae, E. coli
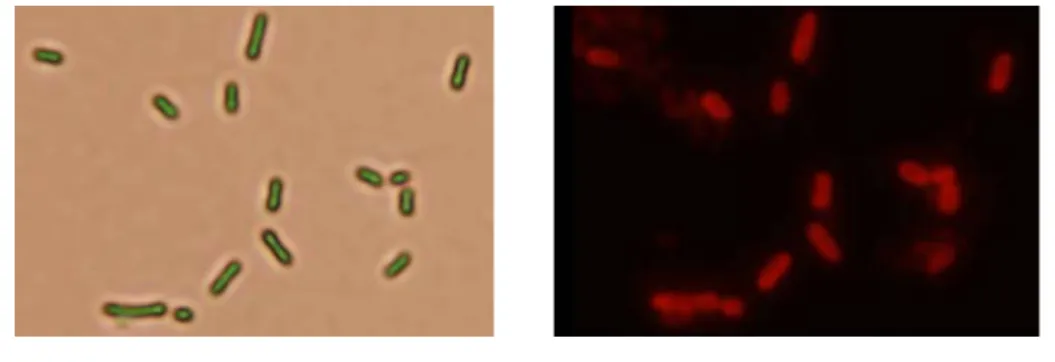
display systems including kpd, kpe, kpf, mrk fimbriae have been obtained and
expression of the fimbriae will be confirmed by analysis using both western blotting hybridization and immunofluorescencent microscopy (Fig. 4). The E. coli
transformants carrying each of the plasmid are being analyzed based on the phenotypic changes including fimbriation activity, autoaggregation, and biofilm
formation.
FIG. 4. Kpe fimbriae on the surface of E. coli JM109 detected using
immunofluorescence microscopy.
(2)-1. As shown in Fig. 1, next to the type 3 fimbriae mrkABCDF gene cluster, orfS and orfM were found following with a fim gene cluster encoding the type 1 fimbriae.
Using either analysis of western blot or immunogold-transmission electronic microscopy (TEM) with anti-MrkA and anti-FimA antisera, the presence of a
cross-talk regulation in controlling the expression of type 1 and type 3 fimbriae was demonstrated. In addition, using lacZ reporter system and gel mobility shift assay, a
regulatory control of the expression of type 3 fimbriae by OrfS/OrfM was determined (manuscript to be submitted).
(2)-2. As shown in Fig. 5, allelic variation of mrkD adhesin of type 3 fimbriae
apparently influence fimbriation of the variants and also their fimbrial activities assessed using cell adherence assay, biofilm formation capability, and collagen
binding assay. Site-directed mutagenesis will be carried out to identify the binding motifs respectively to type IV- and type V-collagen, and to fibronectin.
FIG. 5. Transmission electron micrographs of the type 3 fimbriae displaying on E.
coli. One drop of bacterial suspension was placed on a carbon-coated copper grid,
and negatively stained with 2 % phosphotungstic acid. (A) E. coli JM109 [pGEM-T], (B) E. coli JM109 [pmrkABC], (C) E. coli JM109 [pmrkABCF], (D) E. coli JM109
[pmrkABCDV1F], (E) E. coli JM109 [pmrkABCDV2F], (F) E. coli JM109
[pmrkABCDV3F],(G) E. coli JM109 [pmrkABCDV4F].
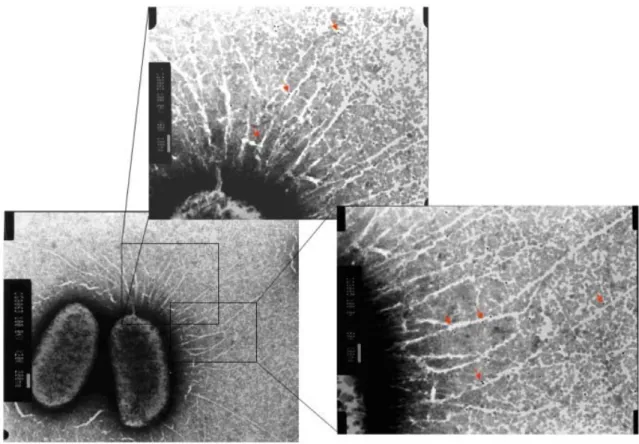
(2)-3. Fimbriation analysis of the E. coli displaying with various recombinants of type 3 fimbriae including mrkABC, mrkABCD, mrkABCF, mrkABCDF using
transmission electronic microscopy (Fig. 6), western blotting analysis, and co-immunoprecipitation, demonstrated that MrkF is a component of type 3 fimbriae.
FIG. 6. Type 3 fimbriae labeled using anti-MrkF immuno-gold. The magnified
V. Literature Cited
1. Allen, B. L., G. –F. Gerlach, and S. Clegg. 1991. Nucleotide sequence and functions of mrk
determinants necessary for expression of type 3 fimbriae in Klebsiella pneumoniae. J. Bacteriol. 173:916-920.
2. Di Martino, P., Y. Bertin, J. P. Girardeau, V. Livrelli, B. Joly, and A. Darfeuille-Michaud.
1995. Molecular characterization and adhesive properties of CF29K, an adhesin of Klebsiella pneumoniae strains involve in nosocomial infections. Infect. Immun. 63:4336-4344.
3. Di Martino, P., V. Livrelli, D. Sirot, B. Joly, and A. Darfeuille-Michaud. 1996. A new
fimbrial antigen harbored by CAZ-5/SHV-4-producing Klebsiella pneumoniae strains involved in nosocomial infections. Infect. Immun. 64:2266-2273.
4. Fader, R. C., K. Gondesen, B. Tolley, D. G. Ritchie, and P. Moller. 1988. Evidence that in
vitro adherence of Klebsiella pneumoniae to ciliated hamster tracheal cells is mediated by type 1 fimbriae. Infect. Immun. 56:3011-3013.
5. Fang CT, C. Y., Shun CT, Chang SC, Wang JT. 2004. A novel virulence gene in Klebsiella
pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med. 199:697-705.
6. Favre-Bonte, S., A. Darfeuille-Michaud, and C. Forestier. 1995. Aggregative adherence of
Klebsiella pneumoniae to human intestine-407 cells. Infect. Immun. 63:1318-1328.
7. Gerlach, G. –F., and S. Clegg. 1988. Characterization of two genes encoding antigenically
distinct type-1 fimbriae of Klebsiella pneumoniae. Gene 64:231-240.
8. Gerlach, G. –F., S. Clegg, and B. L. Allen. 1989. Identification and characterization of the
genes encoding the type 3 and type 1 fimbrial adhesins of Klebsiella pneumoniae. J. Bacteriol. 171:1261-1270.
9. Gerlach, G. B., L. Allen, and S. Clegg. 1989. Type 3 fimbriae among Enterobacteriaceae and
the ability of spermidine to inhibit MR/K hemagglutination. Infect. Immun. 57:219-224. 10. Holden, N.J., Uhlin, B.E., and Gally, D.L. 2001. PapB paralogues and their effect on the
phase variation of type 1 fimbriae in Escherichia coli. Mol Microbiol 42: 319–330.
11. Humphries AD, Raffatellu M, Winter S, Weening EH, Kingsley RA, Droleskey R, Zhang S,
Figueiredo J, Khare S, Nunes J, Adams LG, Tsolis RM, Baumler AJ. 2003. The use of flow
cytometry to detect expression of subunits encoded by 11 Salmonella enterica serotype Typhimurium fimbrial operons. Mol Microbiol. Jun; 48 (5):1357-76.
12. Johnson, D.E., Lockatell, C.V., Russell, R.G., Hebel, J.R., Island, M.D., Stapleton, A., et al. 1998. Comparison of Escherichia coli strains recovered from human cystitis and pyelonephritis infections in transurethrally challenged mice. Infect Immun 66: 3059–3065.
13. Klemm P, B. J. Jorgensen, van Die I, H. de Ree, and H. Bergmans. 1985. The fim genes responsible for synthesis of type 1 fimbriae in Escherichia coli, cloning and genetic
organization. Mol. Gen. Genet. 199:410-414.
14. Klemm P. 1986. Two regulatory fim genes, fimB and fimE, control the phase variation of type 1 fimbriae in Escherichia coli. EMBO J. 5:1389-1393.
15. Klemm P. and G. Christiansen. 1987. Three fim genes required for the regulation of length and mediation of adhesion of Escherichia coli type 1 fimbriae. Mol. Gen. Genet. 2.8:439-445. 16. Schurtz Sebghate, T. A., T. K. Korhonen, D. B. Kornick, and S. Clegg. 1998.
Characterization of the type 3 fimbrial adhesins of Klebsiella strains. Infect. Immun. 66:2887-2894.
17. Soto GE, Hultgren SJ. 1999. Bacterial adhesins: common themes and variations in architecture and assembly. J Bacteriol. Feb; 181(4):1059-71.
18. Tarkkanen, A. M., R. Virkola, S. Clegg, and T. K. Korhonen. 1997. Binding of the type 3 fimbriae of Klebsiella pneumoniae to human endothelial and urinary bladder cells. Infect. Immun. 65:1546-1549.
19. Townsend SM, Kramer NE, Edwards R, Baker S, Hamlin N, Simmonds M, Stevens K,
Maloy S, Parkhill J, Dougan G, Baumler AJ. 2001. Salmonella enterica serovar Typhi
possesses a unique repertoire of fimbrial gene sequences. Infect Immun. May; 69(5):2894-901. 20. van der Velden AW, Baumler AJ, Tsolis RM, Heffron F. 1998. Multiple fimbrial adhesins are
required for full virulence of Salmonella typhimurium in mice. Infect Immun. Jun; 66(6):2803-8.
21. Xia, Y., D. Gally, K. Forsman-Semb, and B. E. Uhlin. 2000. Regulatory cross-talk between adhesin operons in Escherichia coli: inhibition of type 1 fimbriae expression by the Pap B protein. EMBO. 19:1450-1457.
22. Zhang, L., Foxman, B., Manning, S.D., Tallman, P., and Marrs, C.F., 2000 Molecular epidemiologic approaches to urinary tract infection gene discovery in uropathogenic Escherichia coli. Infect Immun 68: 2009–2015.
VI. Self-Assessment
We have carried out the experiments as planned and several manuscripts (listed as the following) are in the final stage to be submitted for publication. In addition,
many interesting findings allow the anticipation of important contributions to unravel the regulation of fimbrial expression at genomic level, the cross-talk
regulatory circuit in-between the nine fimbrial gene clusters, and also their roles in infections, which hold promise to provide intervening targets for antimicrobial
agents.The manuscripts to be submitted include:
1. Chien-Chin Wu, Ying-Jung Huang, Chang-Phone Fung, and Hwei-Ling
Peng, Fimbrial gene cluster analysis for characterization of Klebsiella
pneumoniae clinical isolates.
2. Ying-Jung Huang and Hwei-Ling Peng, Regulation of type 3 fimbriae in
Klebsiella pneumoniae CG43.
3. Ying-Jung Huang, Chien-Chin Wu, Mei-Chen Chen, Chang-Phone Fung,
and Hwei-Ling Peng, Comparative analysis of the type 3 fimbriae with
different mrkD adhesin alleles.
4. Hsin-Wei Liao, Ying-Jung Huang, Chien-Chen Wu, and Hwei-Ling Peng. MrkF is a component of type 3 fimbriae.