半胱胺酸66與232協同調控氧化還原反應性酚亞硫酸基轉移酵素之催化作用
58
0
0
全文
(2) 半胱胺酸 66 與 232 協同調控氧化還原反應性酚亞硫酸基轉移酵素之 催化作用. Cys66 and Cys232 Synergistically Regulate the Catalysis of Redox-Responsive Phenol Sulfotransferase 研 究 生:林志衡. Student: Chih-Heng Lin. 指導教授:楊裕雄 教授. Advisor: Prof. Yuh-Shyong Yang 國 立 交 通 大 學 生化工程研究所 碩 士 論 文 A Thesis. Submitted to Institute of Biochemical Engineering College of Biological Science and Technology National Chiao Tung University in partial Fulfillment of the Requirements for the Degree of Master in Biochemical Engineering July 2006 Hsinchu, Taiwan, Republic of China. 中華民國九十五年七月.
(3) 半胱胺酸 66 與 232 協同調控氧化還原反應性 酚亞硫酸基轉移酵素之催化作用 研究生:林志衡. 指導教授:楊裕雄 教授 國立交通大學生化工程研究所碩士班 摘 要. 氧化還原反應為轉譯後修飾的機制之ㄧ,其可造成蛋白質構型上的改變而影響酵素的 催化效率。大鼠酚亞硫酸基轉移酵素(rat phenol sulfotransferase, rPST 或 rSULT1A1)於不同 的氧化還原環境下,存在著重要的變異於反應催化效率、核苷酸鍵結親和力與反應受質的 專一性。然而,我們發現目前在亞硫酸基轉移酵素(ST)家族當中,唯獨大鼠酚亞硫酸基轉 移酵素具有被氧化還原所調控的特性,其中半胱胺酸(cysteine) 232 僅存在於大鼠酚亞硫酸 基轉移酵素上,並且可能與半胱胺酸 66 形成分子內的雙硫鍵。由已知的結晶結構與計算模 擬的酚亞硫酸基轉移酵素而言,我們提出由大鼠酚亞硫酸基轉移酵素第 62 至 68 的胺基酸 序列所組成的可變動環圈(flexible loop),可能在氧化還原調控上扮演極重要的角色。更進 一步地說明於大鼠酚亞硫酸基轉移酵素可變動環圈上的半胱胺酸 66 可能與半胱胺酸 232 形 成雙硫鍵,而導致酵素活性的改變。經由了解氧化還原調控機制,以及利用蛋白質工程等 技術進而製備出可抵抗氧化影響的突變大鼠酚亞硫酸基轉移酵素,與可以於氧化還原環境 下被調控的突變人類酚亞硫酸基轉移酵素與醇亞硫酸基轉移酵素來印證我們的假說。最 後,初步證實在氧化還原的狀態下,半胱胺酸 66 與 232 於共同調節大鼠酚亞硫酸基轉移酵 素的催化反應上皆是必要的。這將是在氧化還原如何調控酵素的研究上,一個相當重要的 里程碑。 i.
(4) Cys66 and Cys232 Synergistically Regulate the Catalysis of Redox-Responsive Phenol Sulfotransferase Student: Chih-Heng Lin. Advisor: Prof. Yuh-Shyong Yang. Institute of Biochemical Engineering, College of Biological Science and Technology, National Chiao Tung University, Hsinchu, Taiwan, ROC.. ABSTRACT The redox effect, one of the post-translational modifications, could modulate conformational change of the protein and regulate its catalytic efficiency. In the varied redox environment, rat phenol sulfotransferase (rPST, or rSULT1A1) exhibited significant alteration in its catalytic efficiency, nucleotide binding affinity, and substrate specificity. To date, it was found that rPST among the cytosolic ST super-family was solely regulated by redox treatment. Particularly, the Cys232 residue, specifically existing in rPST, has been verified binding to Cys66 by a formation of an intra-molecular disulfide bond. Based on the known crystal structures and computational modeling of STs, we proposed that the flexible loop from amino acid residues 62 to 68 of rPST was crucially related to redox circumambience. We further recommended that Cys66 in the flexible loop formed a disulfide bond with Cys232 resulting in an alteration of its enzyme activity. With the understanding in redox regulation mechanism, rPST mutants were created to resist oxidation, and both hDHEA-ST and hP-PST mutants, which wild types were independent on redox effect, were created to be under redox regulation by protein engineering to confirm our hypothesis. We concluded that Cys66 and Cys232 were both essential to synergistically regulate the rPST catalysis under redox conditions. This was a significant advance for understanding the redox regulation of cytosolic STs.. ii.
(5) Acknowledgement 時光匆匆,回首過去,LEPE 依然是首選。在領航超人 楊裕雄教授的潛移默化, 以及眾多同仁們的護航之下,兩年的旅程終究劃下完美的句點。首要感謝與學習的是深 知灼見的 楊教授,自主管理的領導優勢,亦師亦友並且不厭其煩的肺腑建言,加上首 屈一指的研究資源,提攜成就了無數的研究人才。 再者,感謝口試蒞臨指導的 毛仁淡院長、陳俊榮老師、冉曉雯老師,您們深入且 整體性的指教建議,使得論文與研究更臻完美。此外,前途光明的莊博與奇翰學長,感 謝您們於實驗、論文、未來規劃上的教授,以及前程似錦的偉迪學長、陸宜、江平等 ST 師傅與前輩們的引領入門;郁吟學姊在分生方面的精細指導。然而,好事多磨,謝 謝莊博、政哲、程允學長提供不同領域的種種看法與經驗談,以及恩仕、小木、青辰、 大晃、嘉蔚、鴻斌學長,靜玟、佩綺、小羿學姊,昱樹、淑婷所給予的諸多意見,加上 怡宏、秉鈞、智棚、淵仁學長,惠菁學姊、文穎、韋汝、美春等等的陪伴。LEPE 有了 各位,變得更加蓬蓽生輝,歷久彌堅。 最後,由衷地感激二十幾年來養育扶持的雙親,沒有您們,一切歸零。然而,隱藏 在背後的幕僚長官們國鋆老大、沙巴老哥您們也絕對功不可沒。俗話說:好酒沉甕底, 心存感激地醉了五年半的陳年好酒,吃了秤砣鐵了心打算沉醉直到生命的電池耗盡。. 光陰似箭,歲月如梭,回首過去,歷歷在目。 甲申之秋,初生之犢,丙戌年夏,初生之鼠。. ~ Thank you very much ~ iii.
(6) Contents Chinese Abstract……………………………...……………………………….………………..i English Abstract………………………………………………………………………….…….ii Acknowledgement…………………………………………………………………………….iii Contents……………………………………………………………………….……………....iv Table Contents………………………………………………………….....................................v Figure Contents………………………………………………………………….………….…vi Abbreviation and Symbol……………………………………………………….……….…...vii Introduction…………………………………………………………….....................................1 Experimental Procedures………………………………………………....................................4 Results…………………………………………………………………………….……………8 Discussion…………………………………………………………….....................................12 Conclusion……………………………………………………………………………………19 References……………………………………………………………………………….…....20 Appendix………………………………………………………………………………..…….41. iv.
(7) Table Contents Table 1. Oilgonucleotides primers used for the generation of ST mutants………...………………..…………....27. Table 2. The nucleotide sequences of mutating targets of STs by DNA sequencing……………………………..28. Table 3. Dissociation constants of PAP and STs………………………………………………………………….29. Table 4. Km (PAP) and Vmax of transfer reaction catalyzed by rat and human PST and their mutants……………30. Table 5. Km (PAPS) and Vmax of physiological reaction catalyzed by rat and human ST and their mutants....…..31. Table 6. Kinetic constants of PSTs that catalyzed transfer reaction in redox conditions....…..………………….32. Table 7. Kinetic constants of STs that catalyzed physiological reaction in redox conditions................................33. v.
(8) Figure Contents Figure 1. Physiological catalyzed by STs………………..…………………………...…………………….……...34. Figure 2. Assay of STs…………………………………………………………....…………………..………..…..35. Figure 3. Multiple sequences alignment of wild type STs………………………………………………….……...36. Figure 4. Molecular homologous modeling structure of rPST by SWISS-MODEL………………….……….…..37. Figure 5. Structural comparison of rPST and hSTs by Combinatorial Extension (CE) method…………………..38. Figure 6. The distance between relative two thiol groups of rPST and hSTs……………………………………...39. Figure 7. Protein electrophoresis analysis of all sulfotransferase after redox treatment…………………………..40. vi.
(9) Abbreviation and Symbol Abbreviation and Symbol. Full name. rPST hP-PST hDHEA-ST ε bis-tris propane Bromphenol blue PAP. Rat phenol sulfotransferase Human P-form phenol sulfotransferase Human dihydroepiandrosterone sulfotransferase Absorption (extinction) coefficient 1,3-bis[tris(hydroxymethyl)methylamino]propane 3’, 3’’, 5’, 5’’-tetrabromophenolsulfonephthalein Adenosine 3’,5’-bisphosphate 3’-phosphoadenosine 5’-phosphate Adenosine 3’-phosphate 5’-phosphosulfate 3’-phosphoadenosine 5’-phosphosulfate. PAPS pNP pNPS MUS MU GSH GSSG TCEP DTT EDTA PMSF SDS TEMED DHEA U A280 Kd Km Vmax. p-nitrophenol p-nitrophenyl sulfate 4-methylumbelliferyl sulfate 4-methylumbelliferone glutathione reduced form glutathione oxidized form Tris (2-carboxyethyl) phosphine Dithiothreitol (Ethylenedinitrilo)tetraacetic acid Phenylmethylsulfonyl fluoride Sodium Dodecyl Sulfate N, N, N’, N’-tetramethylethylene diamine Dehydroepiandrosterone Units; μmole/min Absorption at 280 nm Dissociation constant Michaelis constant Maximum velocity. vii.
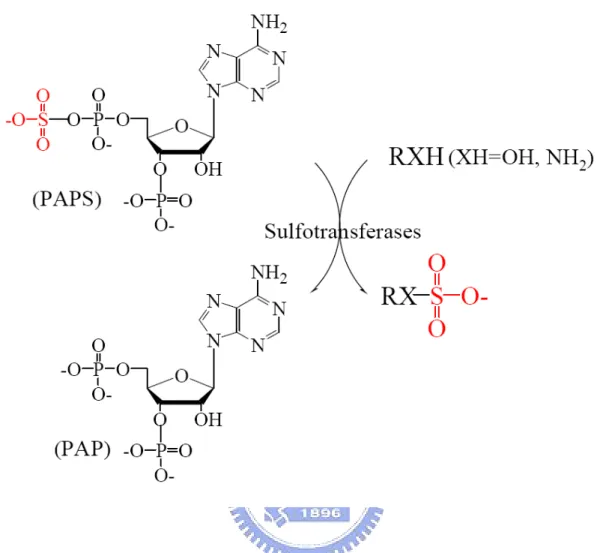
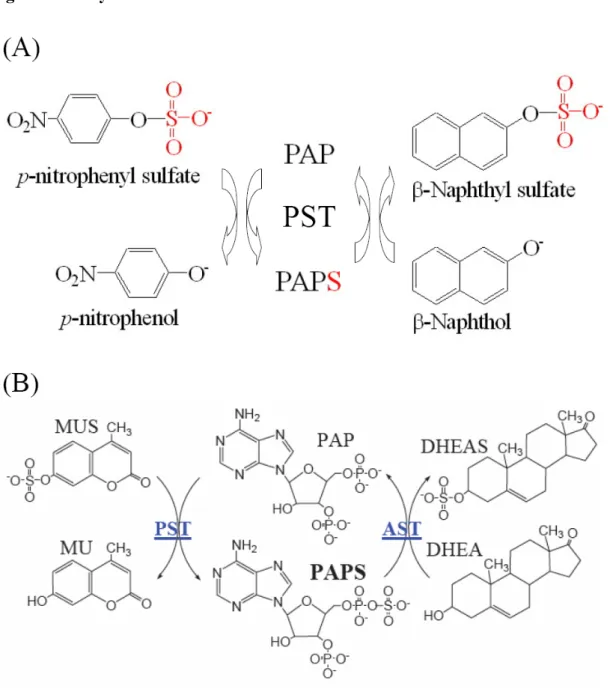
(10) Introduction The sulfonation (also known as sulfurylation) of biomolecules has long been known to take place in a variety of organisms, from prokaryotes to multi-cellular species, and new biological functions continue to be uncovered in connection with this important transformation (Chapman et al., 2004). Sulfotransferases (STs) [EC 2.8.2.X], which catalyze sulfuryl group transfer between a nucleotide and a variety of nucleophiles, are responsible for all the known biological sulfations. The nucleotide, 3’-phosphoadenosine 5’-phosphosulfate (PAPS), is a biologically active form of inorganic sulfate that serves as the universal sulfuryl donor in various biological processes catalyzed by STs as shown in Figure 1. Macromolecular substrates such as proteins and polysaccharides are metabolized by membrane-bound STs. Small molecules like xenobiotics or endogenous compounds including hormones and neurotransmitters are metabolized by cytosolic STs. STs have been classified into several subfamilies according to the degree of the similarities of the deduced amino acid sequences (Nagata and Yamazoe, 2000; Weinshilboum et al., 1997) or substrate specificities (Guo et al., 1994). Recent studies have implicated the STs in a number of disease states including various forms of cancer (Perera, 1997), entry of the herpes virus (Shukla et al., 1999) or HIV (Cormier et al., 2000; Farzan et al., 1999), and chronic inflammation (Kansas, 1996). They are novel therapeutic targets to discover new drugs (Hemmerich, 2001). Phenol sulfotransferases (PSTs) [EC 2.8.2.9] are classified as phase II drug- or xenobiotics-metabolizing enzymes that catalyze reactants to produce excreted hydrophilic products, and play important roles in detoxication. PST (or SULT1A1) have been cloned from rat liver and expressed in Escherichia coli (Chen et al., 1992). During expression of the clone in E. coli, the α (PAP inside)- and β (PAP-free)-forms of the ST are produced and both active. The 1.
(11) expression ratio of the two forms is dependent on cell culture condition (Yang et al., 1998). The two forms are separable from each other by PAP-agarose chromatography (Su and Yang, 2003; Lin and Yang, 2000). Only the β-form can carry out physiological reaction, but both two forms catalyze transfer reaction containing 3’-phosphoadenosine 5’-phosphate (PAP), as shown in Figure 1 and 2 (Yang et al., 1996). Redox and nucleotide binding have been shown to regulate catalytic activity of rPST and are responsible for the complete inter-conversion of two enzyme forms (Su and Yang, 2003). Two conserved motifs that are important for nucleotide (PAP/PAPS) binding in the sulfotransferase family are found (Gamage et al., 2003; Pedersen et al., 2002; Rehse et al., 2002; Pakhomova et al., 2001; Pedersen et al., 2000; Kakuta et al., 1999; Kakuta et al., 1997; Bidwell et al., 1996). The 5’-phosphosulfate binding (PSB) loop, 41-TYPKSGT-47 of rPST, interacts with 5’-phosphate of PAP or 5’-phosphosulfate of PAPS. Another 3’-phosphate binding (PB) loop, which interacts with 3’-phosphate of the nucleotide, is the amino acid sequences 253-RKGXXGDNK-261. The modulation of enzymatic activity is of great importance in biology. The non-equilibrium nature of biological systems necessitates temporal or spatial suppression or enhancement of specific biochemical pathways according to cellular metabolism or exogenous signals. For example, conformational regulation by phosphorylation is a recurring theme in cellular signal transduction pathways. Such in vivo regulation through conformational change inspired us to devise a new “on-off” switch. The oxidation of naturally occurring adjacent cysteine residues can impair protein function. Wild type rPST contains five cysteine residues in each monomer of the homo-dimeric protein (Yang et al., 1996). Previous study confirmed that a highly conserved Cys66 is important for the regulation of rPST catalytic activity, pH optima and substrate specificity through oxidation. The effect of each cysteine on the activity of rPST in different 2.
(12) redox conditions has been demonstrated with a single cysteine mutant (Marshall et al., 2000; Marshall et al., 1997). Only rPST in the cytosolic ST super-family was found to be regulated by redox treatment. Among them, only rPST had Cys232 residue that has been rarified to bind to Cys66 forming the intra-molecular disulfide bond. Here, we proposed Cys66 and Cys232 are both essential to synergistically regulate the rPST catalysis under redox conditions. Using the techniques of protein engineering, single mutant of either Cys66 or Cys232 for rPST was individual established to study the effect of redox on the catalytic characteristics. Furthermore, hP-PST and hDHEA-ST model, parallel supporting our hypothesis, was also constructed trying to understand the redox regulation mechanism of the enzymes.. 3.
(13) Experimental Procedures Materials. p-nitrophenyl sulfate (pNPS), p-nitrophenol (pNP), 3’-phosphoadenosine 5’-phosphate agarose gel (PAP-agarose), and 3’-phosphoadenosine 5’-phosphate (adenosine 3’, 5’-diphosphate; PAP), 3’-phosphoadenosine 5’-phosphosulfate (PAPS), glutathione oxidized form (GSSG) were purchased from Sigma (USA). Tris (2-carboxyethyl) phosphine (TCEP) was obtained from Pierce (USA). HiTrap™ desalting column was obtained from Amersham Pharmacia Biotech Asia Pacific (Hong Kong). All other chemicals were obtained commercially at the highest purity possible.. Computational analysis. ST sequences were obtained from National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/). The structures of hP-PST (or hSULT1A1) and hDHEA-ST (or hSULT2A1) have been solved by x-ray crystal diffraction and can be obtained from Protein Data Bank (PDB, http://www.rcsb.org/pdb/). The PDB IDs are 1LS6 and 1EFH, respectively. The molecular modeling of rPST structure was done at SWISS-MODEL (http://www.expasy.org/swissmod/SWISS-MODEL.html), and the first approach mode was run (Kakuta et al., 1998). The target sequence, rPST, was used hP-PST as template. The full amino acid sequences for known crystal structure STs were aligned using the CLUSTALW v1.8 program at Protein Information Resource (PIR, http://www-nbrf.georgetown.edu/pirwww/search/multaln. html), and protein structures were compared and aligned using the Combinatorial Extension (CE) method (http://cl.sdsc.edu/ce.html).. 4.
(14) Site-directed mutagenesis of the cDNA encoding STs. Site-directed mutagenesis was performed with PfuTurbo® DNA polymerase using QuickChange (Stratagene, La Jolla, CA). All primers for mutagenesis were purchased from Mission Biotech Co., Ltd. (Taiwan). Wild-type rPST and human ST cDNA incorporated in the pET-3c and pGEX-2TK expression vector were used as templates in conjunction with specific mutagenic primers, respectively. Mutated cDNA sequences were confirmed using an ABI Prism 377 DNA sequencer (Applied Biosystems, Foster City, CA) following the standard protocol.. Preparation of the enzymes. Recombinant wild type and mutant ST clones were transformed into Escherichia coli BL21 (DE3). Detail methods were described previously (Chen et al., 2005; Marshall et al., 1998; Yang et al., 1996). STs were extracted by sonication, and purified by DEAE-Sephacel chromatography and GST gene fusion system (Amersham Pharmacia Biotech, Hong Kong). The two forms of ST were separated by PAP-agarose chromatography as described previously (Su and Yang, 2003). The purity of homogeneous enzymes was more than 95% according to SDS-PAGE.. Dissociation constants of PAP and STs. Dissociation constant was determined by fluorescence spectrophotometer as described previously (Lin and Yang, 2000). An aliquot amount of PAP was added into the solution contain ST (0.06 or 1 μM), 50 mM phosphate buffer at pH 7.0. The decrease in intrinsic fluorescence of protein was measured at 340 nm upon excitation at 280 nm and 25°C with a Hitachi spectrofluorescence (F-4500, Japan). The Kd was determined using nonlinear regression by SigmaPlot 2001, V7.0 and Enzyme Kinetics Module, V1.1 (SPSS Inc., Chicago, IL). 5.
(15) Enzymatic assay. The activity of wild-type and mutant hDHEA-ST was determined from the fluorescence increase of MU at 450 nm upon excitation at 360 nm based on a coupled-enzyme assay method, as shown in Figure 2 (Chen et al., 2005). The reaction mixture consisted of 100 mM potassium phosphate buffer at pH 7.0, 20 μM PAPS, 4 mM MUS, 3.2 mU rPST mutant K65E/R68G. Additionally, PST assay was carried out via the change of absorbency at 400 nm due to elimination of free p-nitrophenol (ε = 10500 M-1cm-1 at pH 7.0) as described previously (Kansas, 1996; Duffel and Jakoby, 1981). Transfer reaction mixture consisted of 1 mM p-nitrophenyl sulfate, 50 μM 2-naphthol, 2 μM PAP and 100 mM bis-tris propane at pH 7.0. This was used to determine total sulfotransferase activity. The α-form activity was determined in the absent of PAP. The β-form activity was the difference between total α-form activity. The spectrophotometric physiological assay followed the decrease in absorbance of 100 μM p-nitrophenol when 300 μM PAPS was the sulfuryl group donor. Rates were linear with time when average absorbance changes at 400 nm of less than 0.025 per minute were followed for 2 minutes.. Determination of kinetic constants. Measurements of the kinetic constants for each substrate were performed by varying the concentrations of one substrate, while keeping the other substrates at a fixed and near saturating concentrations. The apparent Km and Vmax were determined using nonlinear regression by SigmaPlot 2001, V7.0 and Enzyme Kinetics Module, V1.1 (SPSS Inc., Chicago, IL).. 6.
(16) Protein electrophoresis analysis of all ST after redox treatment by SDS-PAGE. All enzymes were passed through a HiTrap™ desalting column to remove small molecules and to exchange buffer (100 mM bis-tris propane at pH 7.0, 1 mM EDTA, 10% glycerol, and 125 mM sucrose) before or after treatment. Reduced and oxidized proteins were incubated with 1 mM reductant TCEP and oxidant GSSG in 25°C for 24 hrs, respectively. Desalting enzyme solutions were diluted with SDS sample buffer without reductant, and subjected to electrophoresis under non-reduced conditions on a 12% Tris-glycine SDS-polyacrylamide gel. The gel was stained with Coomassie Brilliant blue following electrophoresis.. 7.
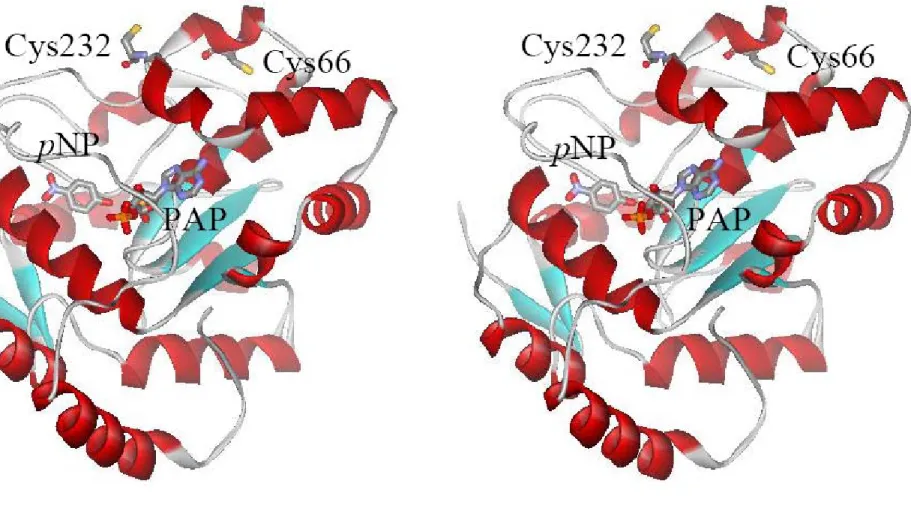
(17) Results Sequence and structural alignment of the STs. There are nine cytosolic ST protein sequences in National Center for Biotechnology Information (NCBI), which are believed to catalyze the transfer of sulfate group (Pakhomova et al., 2001). Following multiple sequence alignment using the CLUSTALW v1.8 program at Protein Information Resource (PIR), Cys66 of rPST and its corresponding residue in other STs is found except for DHEA-ST (SULT2A) that contains an isoleucine (Figure 3). On the contrary, Cys232 only existed in rPST, the corresponding residue in other STs is proline in hPSTs (SULT1A), ESTs (SULT1E), and lysine in hDHEA-STs (Figure 3). To create human ST modeling experiment, we might find out the relative residues of the models (hP-PST and hDHEA-ST) which corresponded to Cys66 and Cys232 of rPST by sequence and structural alignment. As above, Cys66 of rPST was conserved with Cys70 of hP-PST (hSULT1A1), and corresponded to Ile66 of hDHEA-ST (hSULT2A1). Cys232 of rPST corresponded to Pro236 and Lys227 of hP-PST and hDHEA-ST, respectively. Because the crystal structure of rPST is not available at present, we employed molecular homologous modeling to mimic rPST structure using hP-PST (PDB code: 1LS6) as template by SWISS-MODEL, as shown in Figure 4. Similar to that of sequence alignment, the conserved residues of the models (hP-PST and hDHEA-ST) corresponding to Cys66 and Cys232 of rPST are identical after the structural comparison by Combinatorial Extension (CE) method, as shown in Figure 5.. Construction, Expression, and purification of recombinant STs. To study the relationship between cysteines of STs and redox treatment, we purchased synthetic oilgonucleotides as primers shown in Table 1, and prepared six mutating clones, rPST mutant C66S, C232S, hP-PST mutant P236C, 8.
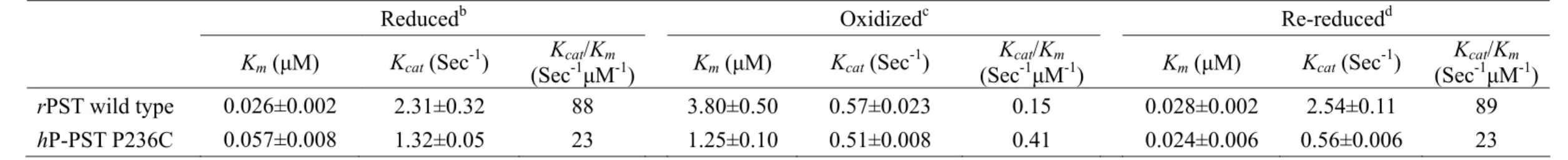
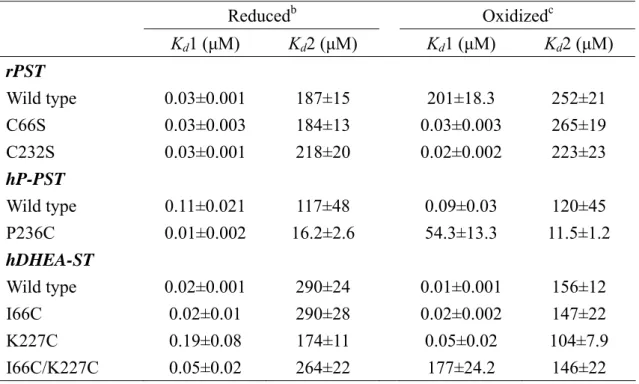
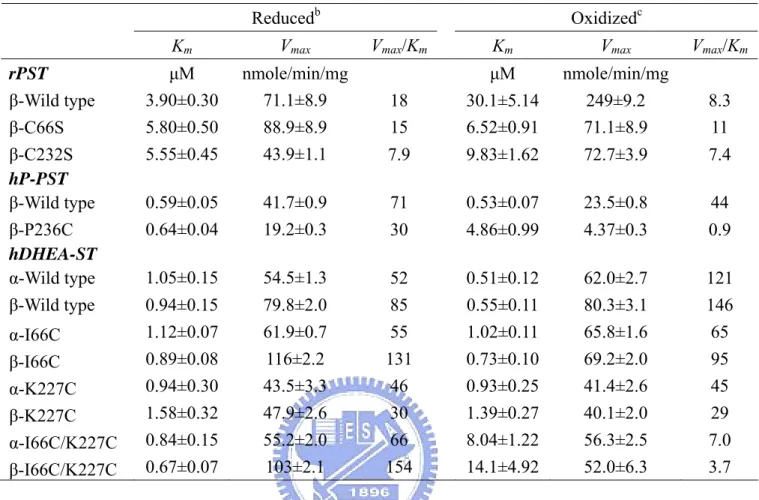
(18) hDHEA-ST mutant I66C, K227C, and I66C/K227C by site-directed mutagenesis. We gained the correct mutant clones and the mutating gene cDNA sequences were confirmed by DNA sequencing, as shown in Table 2. The recombinant wild type and mutant ST clones were over-expressed in E. coli strain BL21 (DE3). All enzymes were purified for the following study and the purity was shown by SDS-PAGE at 34-kDa to at least 95% homogeneity, as shown in Figure 7 (Line 1, 3, 5, 7, 9, 12, 14, 16, and 18).. Effect of redox on the dissociation constants (Kd) of PAP and STs. Table 3 indicated that Kd1 of PAP and rPST wild type (β-form) increased four orders of magnitude in the presence of oxidant (GSSG) rather than reductant (TCEP). The Kd1 and Kd2 of the both rPST mutants C66S and C232S were independent of redox condition while using TCEP and GSSG for reductant and oxidant, respectively. Only the two enzymes, hP-PST mutant P236C and hDHEA-ST mutant I66C/K227C, exhibited similar tendency with rPST wild type where Cys66 and Cys232 co-existed through oxidation. Also the dissociation constants in hP-PST wild type, hDHEA-ST wild type, I66C, and K227C were independent on the oxidization treatment.. Redox effect on the catalytic properties of STs. Activity and kinetic constants of rPST wild type and hP-PST mutant P236C for transfer reaction varied with the redox condition, as shown in Table 4. The Km value, in terms of the concentration of PAP, was observed as the most significant index of enzyme activity under redox condition. After the oxidation treatment, the β-form of rPST wild type and hP-PST mutant P236C exhibited 150- and 20-fold increase, 9.
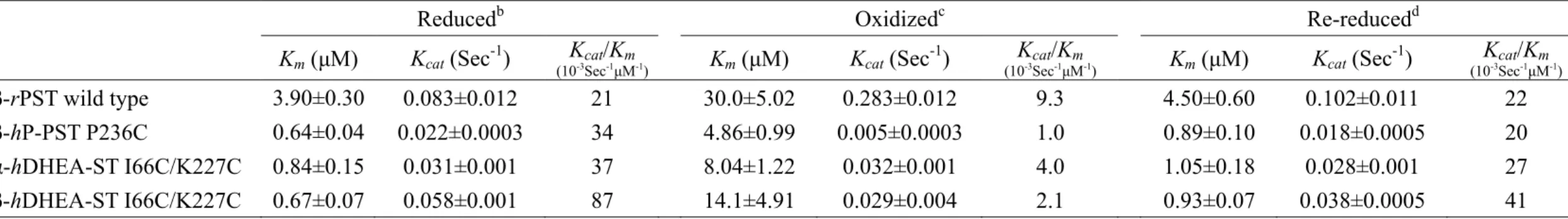
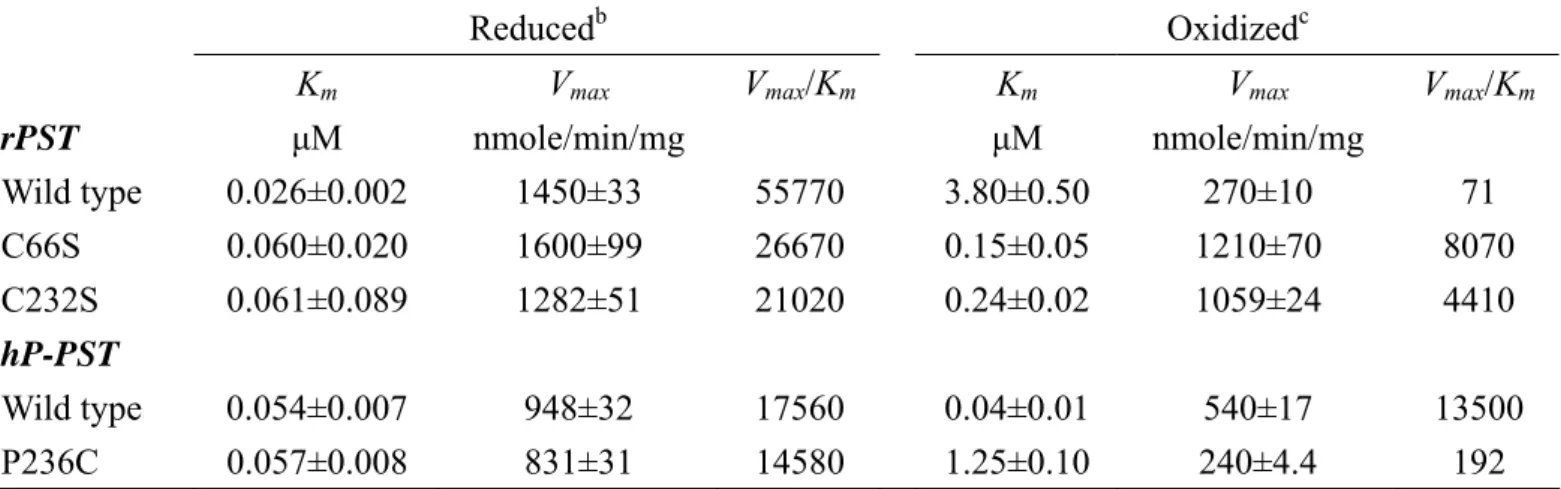
(19) respectively, rather than the corresponding reduced form. Furthermore, the catalytic efficiency (Vmax/Km) in oxidation was 790- and 75-fold lower in rPST wild type and hP-PST mutant P236C than reduction, respectively. The rPST mutants C66S, C232S, and hP-PST wild type that were only one relative cysteine alone were much less different in the redox effect. In addition, Km of PAPS for the physiological reaction using nucleotide (PAPS) as a substrate increased 8-fold under oxidation condition with β-form of wild type rPST rather than reduction, as shown in Table 5, and significant increase of Km was observed for hP-PST mutant P236C (8-fold) and hDHEA-ST mutant I66C/K227C including α- and β-form that were 10- and 20-fold greater in oxidation, respectively, rather than reduction. The Km of PAPS of hP-PST wild type, hDHEA-ST wild type and mutants (I66C and K227C), and rPST mutants (C66S and C232S) without both relative Cys66 and Cys232 weren’t affected by GSSG treatment.. Variation of STs catalytic activity in redox condition. According to the distinct variation of catalytic efficiency and kinetic constants for transfer reaction were also observed for rPST wild type and hP-PST mutant P236C in reduced, oxidized, and re-reduced environments, as shown in Table 6. The overall catalytic efficiency (kcat/Km) of rPST wild type and hP-PST mutant P236C was 590- and 56-fold lower in oxidation, respectively, rather than reduction. All activities of oxidized PSTs were recovered after reduced with TCEP (1mM) at 25℃ for six hours. The kcat and Km of oxidation-sensitive enzymes could be recovered following reduction of the oxidized enzyme. Similar to the results of physiological reaction, as shown in Table 7, the kinetic constants (PAPS) of all oxidization-responsive enzymes including β-rPST wild type, β-hP-PST mutant P236C, and both form of hDHEA-ST mutant I66C/K227C also recurred in re-reduced condition. 10.
(20) Protein electrophoresis analysis of all STs in redox conditions. The electrophoresis patterns of both oxidation-sensitive and resistant proteins after the redox treatment were shown in Figure 7. Relative to the pattern of reduced form, one more evolved band was demonstrated in rPST wild type, hP-PST P236C, and hDHEA-ST I66C/K227C depicted the formation of intra-molecular disulfide bond after incubation of oxidant GSSG. The conserved cysteine residues were found in all of these enzymes in above, thereby showing one more evolved band, whereas, oxidation-resistant proteins, consisting of only one relative or no cysteine in these conserved residues, exhibited single band only in both reduced and oxidized condition.. 11.
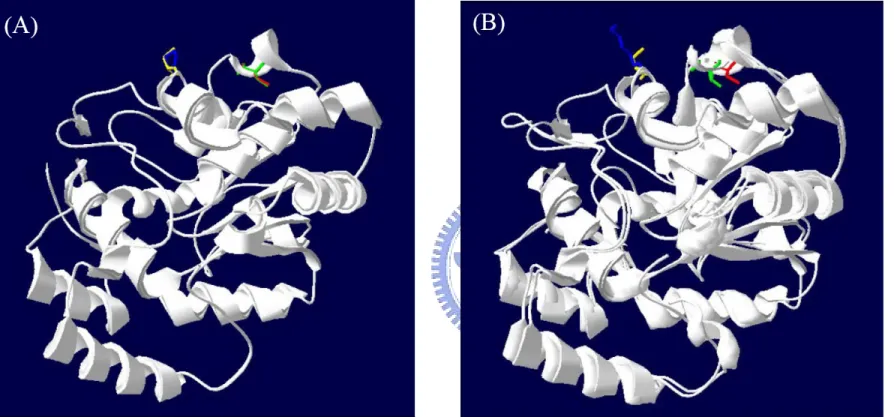
(21) Discussion The formation of intra-molecular disulfide bond between adjacent thiol groups of cysteines. In this study, mutation of cysteine residue was proposed affecting not only the chemical property of the amino acid side chain but also the structure of the enzyme. The disulfide mapping of the tryptic peptides derived after incubation with oxidant (GSSG) confirmed that the event occurring for the wild type rPST at the time of maximum activity consisted of the formation of the intra-molecular disulfide bond between Cys66 and Cys232 (Marshall et al., 1997). By investigating rPST molecular modeling structure (Figure 4) using hP-PST or hSULT1A1 (PDB code: 1LS6) as template, and their amino acid sequence identity was 79.5%; the distance between two thiol groups of Cys66 and Cys232 is 12.8 Å, whereas molecular modeling structure showed 15.1 Å between Cys283 and Cys289 in rPST, as shown in Figure 6. The distance of two thiol groups between Cys82 to other four cysteine residues was over 20 Å, it is extremely hard to formed intra-molecular disulfide bond unless the protein existed with non-native conformation. This indicated Cys66-Cys232 and Cys283-Cys289 formed disulfide bond in oxidative environment reasonably. In Figure 7, the oxidized proteins with cysteine cross-linking exhibited another band compared with reduced forms through SDS-PAGE. According to the known human ST crystal structure, the distance between two thiol groups of Cys70 and Cys236 in hP-PST mutant P236C was 12.8 Å, and an approximated value was observed in hDHEA-ST mutant I66C/K227C (11.8 Å between Cys66 and Cys227), as shown in Figure 6. These results pointed out that the corresponding two cysteine residues were the only route for the formation of intra-molecular disulfide bond to affect the protein structure. Redox-sensitive transcription factor, E. coli OxyR, 12.
(22) was activated by the conformational change due to the formation of disulfide bond (Georgiou, 2002; Choi et al., 2001; Zheng et al., 1998). This phenomenon was similar to the activation of the wild type rPST while forming the disulfide bond between Cys66 and Cys232.. The oxidation affected the nucleotide binding of the enzyme by a conformational change. It has been shown that oxidation of rPST was critically important for the function of the enzyme, and the reversible conformational change was responsible for the regulation of rPST activity dependent on redox condition (Su and Yang, 2003; Marshall et al., 1997). The dissociation constant Kd of PAP thus directly affected the catalyzed reactions of the enzymes. The Kd obtained under reduced and oxidized condition were shown in Table 3. Compared to the reduced condition, the significantly higher Kd about four order of magnitude for three oxidation-sensitive enzymes including rPST wild type, hP-PST mutant P236C, and hDHEA-ST mutant I66C/K227C in oxidation indicated that the presence of both Cys66 and Cys232 were required. However, the Kd of enzymes without conserved cysteine residue (hDHEA-ST wild type) and only one, e.g. either Cys66 or Cys232 shown in rPST only, including rPST mutant C66S, C232S, hP-PST wild type, hDHEA-ST mutant I66C, and K227C were almost identical in the redox conditions. We furthermore compared several kinetic constants (Kd, Km, and Vmax) of either transfer or physiological reaction of the three STs including wild type and mutants in different redox condition. Similar characteristics were observed indicating that the binding of nucleotides (PAP/PAPS) was affected after the oxidant (GSSG) treatment. Alignment results of sequences and structures indicated (Figure 3 and 5), the fragment, from Lys62 to Arg68, formed a postulated flexible loop. Previous studies indicated that the fragment of rPST, from residues 62 to 68, was difficulty resolved in several crystal structure of ST that interpreted the flexibility of this 13.
(23) fragment (Kwon et al., 2001; Pakhomova et al., 2001; Lin and Yang, 2000; Marshall et al., 1997). The flexible loop, residues 62 to 68, and Cys232 were adjacent to nucleotide binding PSB loop (41-TYPKSGT-47) and PB loop (253-RKGXXGDNK-261), respectively. When Cys66 and Cys232 formed intra-molecular disulfide bond though oxidation, the shift of the postulated flexible loop involved conformational alteration of PSB and PB loops, thus further nucleotides (PAP/PAPS) binding or dissociating was affected. Redox effect on enzymatic catalysis of transfer reaction was strongly dependent on the presence of both cysteines in PSTs, as shown in Table 4. The Vmax for the transfer reaction employing PAP as a cofactor reduced significantly in oxidative environment for rPST wild type and hP-PST mutant P236C, the only two enzymes containing both Cys66 and Cys232. The conformational change of enzymes through oxidation mainly altered nucleotide (PAP) binding, leading to the significant increase of Km and reduction of sulfate group transfer. Whereas, the kinetic constants of transfer reactions in the enzymes that only had a single cysteine residue including rPST mutant C66S, C232S, and hP-PST wild type were not affected. In other words, the oxidation of adjacent cysteine residues destabilized the protein conformation. The behavior was ever observed in human ribonuclease inhibitor (hRI) where the oxidation of cysteine residues inactivated the protein. Four of 32 cysteine residues in hRI, Cys94, Cys95, Cys328 and Cys329, belonged to two pairs of adjacent cysteine residues. When either pair was replaced with alanine, the resistance of this protein to oxidation was enhanced (Kim et al., 1999).. Both α- and β-forms of hDHEA-ST catalyzed physiological reaction. Theoretically either physiological or reverse physiological reaction should be faster than the transfer reaction. They had only one step, but transfer reaction had two. Whether the reaction was proceed through a bi 14.
(24) bi or ordered mechanism remains controversial. The mechanism of ST was random rapid equilibrium bi bi kinetic mechanism in physiological reaction (Yang et al., 1998; Duffel and Jokoby, 1981), and it was ping-pong bi bi kinetic mechanism in transfer reaction (Kwon et al., 2001; Yang et al., 1998). Only the β-form PSTs could carry out physiological reaction, but both α- and β-forms could finish transfer reaction containing PAP (Yang et al., 1996). The difference between two forms PST was that α-form contained un-releasable PAP, but β-form did not (Su and Yang, 2003). However, we also utilized PAP-agarose chromatography to separate each other of hDHEA-ST including wild type and mutants that could catalyze physiological reaction as shown in Table 5. Therefore, such difference was able to clarify via the diversities of protein sequences and structures between phenol and dihydroepiandrosterone (alcohol) STs.. Shift of the flexible loop opened another passage for the nucleotide in rPST after oxidation. The difference in kinetic mechanisms of physiological and the transfer reaction involved the release of PAP/PAPS during the course of sulfate group transfer (Marshall et al., 2000; Dajani et al., 1999; Yang et al., 1996). In physiological reaction, the product, PAP, was released completing a turnover of a sulfate group transfer (Yang et al., 1996). The PAPS might be not release in transfer reaction. As compared with the kinetic constants of the physiological reaction, there were variable in the Vmax of oxidation-sensitive enzymes including β-rPST wild type, β-hP-PST mutant P236C, and both form of hDHEA-ST mutant I66C/K227C which possessed both relative cysteine residues, as shown in Tables 5. Only oxidized rPST increased the Vmax of the physiological reaction compared with human ST models. The physiological reaction of ST required the release of nucleotides (PAPS/PAP) as a substrate or product to continue for the next run of catalysis (Yang et al., 1998; Yang et al., 1996). 15.
(25) Oxidation following reduction of rPST is required to completely interconvert the two forms of rPST (Su and Yang, 2003). Based on the mechanism of the reversible conversion of the redox forms, we proposed that this flexible loop was like a lid and could alter the entrance and release of the nucleotide. The flexible loop hid the crevice of rPST in reducing environment. When the Cys66 and Cys232 conjugated with disulfide bond, this cover opened at oxidative stage and nucleotide could be release without hindering, consequently the physiological activity of wild type rPST increased. We assumed that conformational change caused by the formed intra-molecular disulfide bond upon oxidation affected the binding of nucleotides as significant increase of the Km in oxidation-responsive enzymes. Furthermore, nucleotide analog was shown to interact with the flexible loop (Zheng et al., 1994), the opening of this region made another passage for nucleotides in and out in merely redox-regulated rPST. On the other hand, the single mutant, either C66S or C232S, could not alter activities in redox environment (Table 5). Spodoptera frugiperda retinol dehydratase catalyzed the conversion of retinol to the retro-retinoid anhydroretinol (Pakhomova et al., 2001). It shared sequence homology with the family of mammalian cytosolic STs and had the activity of STs. There was 32-amino acid insertion, from Met110 to Tyr141, was the major retinol binding site. It formed a snugly fitting cap for the β-ionone ring of the substrate and as a lid from crystal structure. It made retinol dehydratase distinct from ST. This was powerful evidence that this loop controlled the pathway of the substrate. The flexible loop, constituted by amino acid residues 62 to 68, controlled nucleotide releasing and affected rPST activity at redox condition at the same phenomenon. However, there might be other regulation mechanisms, e.g. different gene expression level (Maiti et al., 2005; Maiti et al., 2004) for human STs by nature. So, the decreased Vmax of human ST 16.
(26) mutant models for the physiological reaction might result from enzyme structural alterations like transfer reaction.. The catalytic characteristics of oxidized enzymes were recovered upon reductant treatment. From our study and previous results (Su and Yang, 2003), we found the rate-limiting step of the PST enzymatic reaction was the association and dissociation of PAP/PAPS. The movement of the fragment did not influence pNPS binding, so the Kd for 2-naphthol and 2-naphthyl sulfate was the same of oxidized rPST (Marshall et al., 1997). The PAP-releasing step was easier, and needed more sulfuryl group donor (PAPS) to reach greater Vmax for oxidized rPST. So the Kd for PAP became larger in oxidized rPST wild type as shown in Table 3, and needed more PAPS for physiological reaction to saturate substrate binding sites reaching Vmax and Km increased reasonably (Table 5). The PAP had more opportunity to leak from binding pocket, and Vmax and Vmax/Km of transfer reaction reduced as shown in Table 4. However, oxidation indeed affected several enzymatic characteristics including nucleotides binding and catalytic activity, whether there were redox regulation in native rPST wild type and artificial human STs by protein engineering or not. The capabilities of oxidized enzymes were recovered, either transfer or physiological reaction as shown in Table 6 and 7. The formed intra-molecular disulfide bond of oxidized enzyme interrupted that shifted postulate flexible loop should return to original position and the transformed structure would be changed to native conformation through reductant (TCEP) treatment. Then the catalytic activity of re-reduced enzyme could revert to primitive one. For example, oxidation of Cys558 and Cys559 in the active site of mercuric reductase abolishes its catalytic activity (Miller et al., 1989). When the disulfide bond is reduced with dithiothreitol (DTT), the enzyme recovers its activity. 17.
(27) The redox regulation in vivo. Cellular redox signaling contributed to the control of cell development, differentiation, growth, death and adaptation, and had been implicated in diverse physiological and pathological processed (Droge, 2002). Two forms rPST were reversible by redox method in vitro (Su and Yang, 2003). The concentration of PAP and glutathione were in μM (Lin and Yang, 1998) and mM range (Senft et al., 2000) in rat liver. The glutathione redox couple was present in mammalian cell in concentrations between 1 and 10 mM, with the reduced (GSH) predominating over the oxidized (GSSG) form (Tietze, 1969). In the resting cell, the ratio of GSH to GSSG exceeded 100, whereas in various models of oxidative stress this ratio was reported to decrease to values between 10 and 1 (Gilbert, 1995). The loop had opportunity as a nano-switch controlling the post-translational modification, nucleotide in or out, by redox signaling in vivo. This research provided further binding information of nucleotide in the redox-responsive condition. The binding site of a protein was usually very selective for particular molecule and with specific function. With the known biological function of ST, the complexity of the binding of ST with a variety of important nucleotides might indicate that regulation of biological function was involved. We have demonstrated that nucleotide binding play important role on the post-translational modification of rPST. Then we proposed that a flexible loop fragment of rPST, from residues 62 to 68, affected nucleotides binding. The mechanism involved a flexible loop function as a lid that controls the in and out of nucleotides. The lid was closed in reducing environment and open in oxidative condition with the formation of an intra-molecular disulfide bond between Cys66 and Cys232.. 18.
(28) Conclusion We utilized molecular homologous modeling to mimic the structure of rPST. By using the sequence alignment and structural comparison, we have described the importance of relative cysteine residues in rPST. Based on the results of the alignments, we create two human sulfotransferase models to verify the proposal by protein engineering. The results of dissociation and kinetic constants illustrate the alteration of nucleotides binding through oxidation. Combining our experimental results with previous literatures, we proposed that the postulated flexible loop fragment from Lys62 to Arg68 of rPST would shift to cause conformational change of the enzyme by the formation of intra-molecular disulfide bond between Cys66 on the loop and Cys232 after oxidation. Furthermore, the capabilities of oxidized enzymes recovered, either in rPST wild type or human sulfotransferase mutant models. Conclusively, redox is a crucial effect to regulate the functional mechanism of the enzymes.. 19.
(29) References 1.. Bidwell, L. M., McManus, M. E., Gaedigk, A., Kakuta, Y., Negishi, M., Pedersen, L., Martin, J. L. (1999) Crystal structure of human catecholamine sulfotransferase, J. Mol. Biol. 293, 521-530.. 2.. Chapman, E., Best, M. D., Hanson, S. R., Wong, C. H. (2004) Sulfotransferases: Structure, Mechanism, Biological Activity, Inhibition, and Synthetic Utility, Angew. Chem. Int. Ed. 43, 3526-3548.. 3.. Chen, W. T., Liu, M. C., and Yang, Y. S. (2005) Fluorometric assay for alcohol sulfotransferase, Anal. Biochem. 339, 54-60.. 4.. Chen, X., Yang, Y. -S., Zheng, Y., Martin, B. M., Duffel, M. W., and Jakoby, W. B. (1992) Tyrosine-ester sulfotransferase from rat liver: bacterial expression and identification, Protein Expr. Purif. 3, 421-426.. 5.. Choi, H., Kim, S., Mukhopadhyay, P., Cho, S., Woo, J., Storz, G., Ryu, S. (2001) Structural basis of the redox switch in the OxyR transcription factor, Cell 105, 103-113.. 6.. Cormier, E. G., Persuh, M., Thompson, D. A., Lin, S. W., Sakmar, T. P., Olson, W. C., Dragic, T. (2000) Specific interaction of CCR5 amino-terminal domain peptides containing sulfotyrosines with HIV-1 envelope glycoprotein gp120, Proc. Natl. Acad. Sci. U.S.A 97, 5762-5767.. 7.. Dajani, R., Cleasby, A., Neu, M., Wonacott, A. T., Jhoti, H., Hood, A. M., Modi, S., Hersey, 20.
(30) A., Taskinen, J., Cooke, R. M., Manchee, G. R., and Coughtrie, M. W. H. (1999) X-ray crystal structure of human dopamine sulfotransferase, SULT1A3. Molecular modeling and quantitative structure-activity relationship analysis demonstrate a molecular basis for sulfotransferase substrate specificity, J. Biol. Chem. 274, 37862-37868. 8.. Droge, W. (2002) Free radicals in the physiological control of cell function, Physiol. Rev. 82, 47-95.. 9.. Duffel, M. W., and Jakoby, W. B. (1981) On the mechanism of aryl sulfotransferase, J. Biol. Chem. 256, 11123-11127.. 10. Farzan, M., Mirzabekov, T., Kolchinsky, P., Wyatt, R., Cayabyab, M., Gerard, N. P., Gerard, C., Sodroski, J., Choe, H. (1999) Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry, Cell 96, 667-676. 11. Gamage, N. U., Duggleby, R. G., Barnett, A. C., Tresillian, M., Latham, C. F., Liyou, N. E., McManus, M. E., Martin, J. L. (2003) Structure of a human carcinogen-converting enzyme, SULT1A1. Structural and kinetic implications of substrate inhibition, J. Biol. Chem. 278, 7655-7662. 12. Georgiou, G. (2002) How to flip the (redox) switch, Cell 111, 607-610. 13. Gilbert, H. F. (1995) Thiol/disulfide exchange equilibria and disulfide bond stability, Methods Enzymol. 251, 8-28.. 21.
(31) 14. Guo, W. -X., Yang, Y. -S., Chen, X., McPhie, P., and Jakoby, W. B. (1994) Changes in substrate specificity of the recombinant form of phenol sulfotransferase IV (tyrosine-ester sulfotransferase), Chem. Biol. Interact. 92, 25-31. 15. Hemmerich, S. (2001) Carbohydrate sulfotransferases: novel therapeutic targets for inflammation, viral infection and cancer, Drug Discov. Today 6, 27-35. 16. Kansas, G. S. (1996) Selectins and their ligands: current concepts and controversies, Blood 88, 3259-3287. 17. Kakuta, Y., Pedersen, L. G., Carter, C. W., Negishi, M., Pedersen, L. C. (1997) Crystal structure of estrogen sulphotransferase, Nat. Struct. Biol. 4, 904-908. 18. Kakuta, Y., Pedersen, L. G., Chae, K., Song, W. C., Leblanc, D., London, R., Carter, C. W., and Negishi, M. (1998) Mouse steroid sulfotransferases: substrate specificity and preliminary X-ray crystallographic analysis, Biochem. Pharm. 55, 313-317. 19. Kakuta, Y., Sueyoshi, T., Negishi, M., Pedersen, L. C. (1999) Crystal structure of the sulfotransferase domain of human heparan sulfate N-deacetylase/ N-sulfotransferase 1, J. Biol. Chem. 274, 10673-10676. 20. Kim, B. M., Schultz, L. W., Raines, R. T. (1999) Variants of ribonuclease inhibitor that resist oxidation, Protein Sci. 8, 430-434.. 22.
(32) 21. Kwon, A. R., Yun, H. J., Choi, E.C. (2001) Kinetic mechanism and identification of the active site tyrosine residue in Enterobacter amnigenus arylsulfate sulfotransferase, Biochem. Biophys. Res. Commun. 285, 526-529. 22. Lin, E. -S., and Yang, Y. -S. (1998) Colorimetric determination of the purity of 3'-phospho adenosine 5'-phosphosulfate and natural abundance of 3'-phospho adenosine 5'- phosphate at picomole quantities, Anal. Biochem. 264, 111-117. 23. Lin, E. -S., and Yang, Y. -S. (2000) Nucleotide binding and sulfation catalyzed by phenol sulfotransferase, Biochem. Biophys. Res. Commun. 271, 818-822. 24. Marshall, A. D., Darbyshire, J. F., Hunter, A. P., McPhie, P., Jakoby, W. B. (1997) Control of activity through oxidative modification at the conserved residue Cys66 of aryl sulfotransferase IV, J. Biol. Chem. 272, 9153-9160. 25. Marshall, A. D., McPhie, P., and Jakoby, W. B. (2000) Redox control of aryl sulfotransferase specificity, Arch. Biochem. Biophys. 382, 95-104. 26. Mattock, P., and Jones, J. G. (1970) Partial purification and properties of an enzyme from rat liver that catalyses the sulphation of L-tyrosyl derivatives, Biochem. J. 116, 797-803. 27. Miller, S. M., Moore, M. J., Massey, V., Williams, C. H. Jr., Distefano, M. D., Ballou, D. P., Walsh, C. T. (1989) Evidence for the participation of Cys558 and Cys559 at the active site of mercuric reductase, Biochemistry 28, 1194-1205.. 23.
(33) 28. Pakhomova, S., Kobayashi, M., Buck, J., Newcomer, M. E. (2001) A helical lid converts a sulfotransferase to a dehydratase, Nat. Struct. Biol. 8, 447-451. 29. Pedersen, L. C., Petrotchenko, E., Shevtsov, S., Negishi, M. (2002) Crystal structure of the human estrogen sulfotransferase-PAPS complex: evidence for catalytic role of Ser137 in the sulfuryl transfer reaction, J. Biol. Chem. 277, 17928-17932. 30. Pedersen, L. C., Petrotchenko, E. V., Negishi, M. (2000) Crystal structure of SULT2A3, human hydroxysteroid sulfotransferase, FEBS Lett. 475, 61-64. 31. Perera, F. P. (1997) Environment and cancer: who are susceptible? Science 278, 1068-1073. 32. Rehse,. P.. H.,. Zhou,. M.,. Lin,. S.. X.. (2002). Crystal. structure. of. human. dehydroepiandrosterone sulphotransferase in complex with substrate, Biochem. J. 364, 165-171. 33. Senft, A. P., Dalton, T. P., Shertzer, H. G. (2000) Determining glutathione and glutathione disulfide using the fluorescence probe o-phthalaldehyde, Anal. Biochem. 280, 80-86. 34. Shukla, D., Liu, J., Blaiklock, P., Shworak, N. W., Bai, X., Esko, J. D., Cohen, G. H., Eisenberg, R. J., Rosenberg, R. D., Spear, P. G. (1999) A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry, Cell 99, 13-22. 35. Maiti, S., Dutta, S. M., Baker, S. M., Zhang, J., Narasaraju, T., Liu, L., Chen, G. (2005) In vivo and In vitro oxidative regulation of rat aryl sulfotransferase IV (AST IV), J. Biochem. Mol. Toxicol. 19, 109-118. 24.
(34) 36. Maiti, S., Grant, S., Baker, S. M., Karanth, S., Pope, C. N., Chen, G. (2004) Stress regulation of sulfotransferases in male rat liver, Biochem. Biophys. Res. Commun. 323, 235-241. 37. Nagata, K., and Yamazoe, Y. (2000) Pharmacogenetics of sulfotransferase, Annu. Rev. Pharmacol. Toxicol. 40, 159-176. 38. Su, T. -M., and Yang, Y. -S. (2003) Mechanism of Posttranslational Regulation of Phenol Sulfotransferase: Expression of two enzyme forms through redox modification and nucleotide binding, Biochemistry 42, 6863-6870. 39. Tietze, F. (1969) Enzymic method for quantitative determination of nanogram amounts of total and oxidized glutathione: applications to mammalian blood and other tissues, Anal Biochem. 27, 502-522. 40. Weinshilboum, R. M., Otterness, D. M., Aksoy, I. A., Wood, T. C., Her, C., and Raftogianis, R. B. (1997) Sulfation and sulfotransferases 1: Sulfotransferase molecular biology: cDNAs and genes, FASEB J. 11, 3-14. 41. Yang, Y. -S., Marshall, A. D., McPhie, P., Guo, W. X., Xie, X., Chen, X., Jakoby, W. B. (1996) Two phenol sulfotransferase species from one cDNA: nature of the differences, Protein Expr. Purif. 8, 423-429. 42. Yang, Y. -S., Tsai, S. -W., Lin, E. -S. (1998) Effects of 3'-phosphoadenosine 5'-phosphate on the activity and folding of phenol sulfotransferase, Chem. Biol. Interact. 109, 129-135.. 25.
(35) 43. Zheng, M., Aslund, F., Storz, G. (1998) Activation of the OxyR transcription factor by reversible disulfide bond formation, Science 279, 1718-1721. 44. Zheng, Y., Bergold, A., Duffel, M. W. (1994) Affinity labeling of aryl sulfotransferase IV. Identification of a peptide sequence at the binding site for 3'-phosphoadenosine-5'phosphosulfate, J. Biol. Chem. 269, 30313-30319.. 26.
(36) Table 1: Oilgonucleotides primers used for the generation of ST mutants Primer rPST C66S. rPST C232S. hP-PST P236C. hDHEA-ST I66C. Sequencesa. Direction Forward. 5'-GCTAGAGAAGAGTGGCCGCGCCCCC-3'. Reverse. 5'-GGGCGCGGCCACTCTTCTCTAGCTTGCC-3'. Forward. 5'-GAAAGAGAACAGCATGACTAACTACAC-3'. Reverse. 5'-AGTTAGTCATGCTGTTCTCTTTCATTTTC-3'. Forward. 5'-GAAGAAGAACTGCATGACCAACTACACC-3'. Reverse. 5'-AGTTGGTCATGCAGTTCTTCTTCATCTC-3'. Forward. 5'-TGCCAAGTGGTGCCAATCTGTGCCCATC-3'. Reverse. 5'-GCACAGATTGGCACCACTTGGCATCCCC-3'. Forward. 5'-GAAAGAAAACTGCATGTCCAATTATTCCC-3'. Reverse. 5'-AATTGGACATGCAGTTTTCTTTCATGCTC-3'. hDHEA-ST K227C. a. Bold and underlined nucleotides indicated the designated positions for mutation.. 27.
(37) Table 2: The nucleotide sequences of mutating targets of STs by DNA sequencing Enzyme. Target amino acid position and nucleotide sequences. rPST Wild type C66S C232S. Cys66 (TGT) Ser66 (AGT) ―. Cys232 (TGC) ― Ser232 (AGC). hP-PST Wild type P236C. Cys70 (TGT) ―. Pro236 (CCT) Cys236 (TGC). hDHEA-ST Wild type. Ile66 (ATC). Lys227 (AAG). I66C K227C I66K/227C. Cys66 (TGC) ― Cys66 (TGC). ― Cys227 (TGC) Cys227 (TGC). The nucleotide sequence was confirmed by DNA sequencing.. 28.
(38) Table 3: Dissociation constants of PAP and STsa Reducedb. Oxidizedc. Kd1 (μM). Kd2 (μM). Kd1 (μM). Kd2 (μM). rPST Wild type C66S C232S. 0.03±0.001 0.03±0.003 0.03±0.001. 187±15 184±13 218±20. 201±18.3 0.03±0.003 0.02±0.002. 252±21 265±19 223±23. hP-PST Wild type P236C. 0.11±0.021 0.01±0.002. 117±48 16.2±2.6. 0.09±0.03 54.3±13.3. 120±45 11.5±1.2. hDHEA-ST Wild type I66C K227C. 0.02±0.001 0.02±0.01 0.19±0.08. 290±24 290±28 174±11. 0.01±0.001 0.02±0.002 0.05±0.02. 156±12 147±22 104±7.9. I66C/K227C. 0.05±0.02. 264±22. 177±24.2. 146±22. a. Dissociation constants were determined by fluorescence as described under Experimental. Procedures. The mean ± standard error was obtained from nonlinear regression program. The β-form of STs (60 nM and 1 μM, dimmer) was used for the determination of Kd1 and Kd2, respectively. b. The STs were passed through a HiTrap™ desalting column to remove small molecules and to. exchange buffer (100 mM bis-tris propane at pH 7.0, 1 mM EDTA, 10% glycerol, and 125 mM sucrose). Desalted STs were incubated in 1 mM TCEP for six hours at 25℃, then passed through HiTrap™ desalting column to remove small molecules. c. Desalted rPST and human STs were incubated in 1 mM GSSG for one and six hours at 25℃,. respectively. Then samples were passed through HiTrap™ desalting column.. 29.
(39) Table 4: Km (PAP) and Vmax of transfer reaction catalyzed by rat and human PST and their mutantsa Reducedb Vmax. rPST Wild type C66S C232S. Km μM 0.026±0.002 0.060±0.020 0.061±0.089. nmole/min/mg 1450±33 1600±99 1282±51. hP-PST Wild type P236C. 0.054±0.007 0.057±0.008. 948±32 831±31. Oxidizedc Vmax/Km. Vmax. Vmax/Km. 55770 26670 21020. Km μM 3.80±0.50 0.15±0.05 0.24±0.02. nmole/min/mg 270±10 1210±70 1059±24. 71 8070 4410. 17560 14580. 0.04±0.01 1.25±0.10. 540±17 240±4.4. 13500 192. a. Enzyme activities were determined as described under experimental procedures. The mean ±. standard error was obtained from nonlinear regression program. b. The β-form PSTs were passed through a HiTrap™ desalting column to remove small. molecules and to exchange buffer (100 mM bis-tris propane at pH 7.0, 1 mM EDTA, 10% glycerol, and 125 mM sucrose). Desalted PSTs were incubated in 1 mM TCEP for six hours at 25℃, then passed through HiTrap™ desalting column to remove small molecules. c. Desalted rPST and hP-PST were incubated in 1 mM GSSG for one and six hours at 25℃,. respectively. Then samples were passed through HiTrap™ desalting column.. 30.
(40) Table 5: Km (PAPS) and Vmax of physiological reaction catalyzed by rat and human ST and their mutantsa Reducedb Km. Vmax. μM 3.90±0.30 5.80±0.50 5.55±0.45. nmole/min/mg 71.1±8.9 88.9±8.9 43.9±1.1. 0.59±0.05 0.64±0.04. Oxidizedc Vmax/Km. Vmax/Km. Km. Vmax. μM. nmole/min/mg. 18 15. 30.1±5.14 6.52±0.91. 249±9.2 71.1±8.9. 8.3 11. 7.9. 9.83±1.62. 72.7±3.9. 7.4. 41.7±0.9 19.2±0.3. 71. 0.53±0.07. 23.5±0.8. 44. 30. 4.86±0.99. 4.37±0.3. 0.9. 0.51±0.12 0.55±0.11. 62.0±2.7 80.3±3.1. 121 146. 1.02±0.11 0.73±0.10 0.93±0.25 1.39±0.27 8.04±1.22 14.1±4.92. 65.8±1.6 69.2±2.0 41.4±2.6 40.1±2.0 56.3±2.5 52.0±6.3. 65 95 45 29 7.0 3.7. rPST β-Wild type β-C66S β-C232S hP-PST β-Wild type β-P236C hDHEA-ST α-Wild type β-Wild type. 1.05±0.15. 54.5±1.3. 0.94±0.15. 79.8±2.0. 52 85. α-I66C β-I66C α-K227C β-K227C α-I66C/K227C β-I66C/K227C. 1.12±0.07 0.89±0.08 0.94±0.30 1.58±0.32 0.84±0.15 0.67±0.07. 61.9±0.7 116±2.2 43.5±3.3 47.9±2.6 55.2±2.0 103±2.1. 55 131 46 30 66 154. a. Enzyme activities were determined as described under experimental procedures. The mean ±. standard error was obtained from nonlinear regression program. b. The STs were passed through a HiTrap™ desalting column to remove small molecules and to. exchange buffer (100 mM bis-tris propane at pH 7.0, 1 mM EDTA, 10% glycerol, and 125 mM sucrose). Desalted STs were incubated in 1 mM TCEP for six hours at 25℃, then passed through HiTrap™ desalting column to remove small molecules. c. Desalted rPST and human STs were incubated in 1 mM GSSG for one and six hours at 25℃,. respectively. Then samples were passed through HiTrap™ desalting column.. 31.
(41) Table 6: Kinetic constants of β-form PSTs that catalyzed transfer reaction in redox conditionsa Reducedb. rPST wild type hP-PST P236C. Km (μM). Kcat (Sec-1). 0.026±0.002 0.057±0.008. 2.31±0.32 1.32±0.05. Oxidizedc Kcat/Km (Sec-1μM-1) 88 23. Km (μM). Kcat (Sec-1). 3.80±0.50 1.25±0.10. 0.57±0.023 0.51±0.008. Re-reducedd Kcat/Km (Sec-1μM-1) 0.15 0.41. Km (μM). Kcat (Sec-1). 0.028±0.002 0.024±0.006. 2.54±0.11 0.56±0.006. Kcat/Km (Sec-1μM-1) 89 23. a. Enzyme activities were determined as described under experimental procedures. The mean ± standard error was obtained from nonlinear. regression program. b. The β-form PSTs were passed through a HiTrap™ desalting column to remove small molecules and to exchange buffer (100 mM bis-tris. propane at pH 7.0, 1 mM EDTA, 10% glycerol, and 125 mM sucrose). Desalted PSTs were incubated with 1 mM TCEP at 25℃ for six hours, then passed through HiTrap™ desalting column to remove small molecules. c. The desalted β-rPST wild type and β-hP-PST mutant P236C were incubated with 1mM GSSG for one and six hours at 25℃, respectively. Then. samples were passed through HiTrap™ desalting column. d. The oxidized PSTs were incubated with 1mM TCEP for six hours at 25℃, then passing through HiTrap™ desalting column.. 32.
(42) Table 7: Kinetic constants of STs that catalyzed physiological reaction in redox conditionsa Reducedb. Oxidizedc. Re-reducedd. Km (μM). Kcat (Sec-1). Kcat/Km (10-3Sec-1μM-1). Km (μM). Kcat (Sec-1). Kcat/Km (10-3Sec-1μM-1). Km (μM). Kcat (Sec-1). Kcat/Km (10-3Sec-1μM-1). β-rPST wild type. 3.90±0.30. 0.083±0.012. 21. 30.0±5.02. 0.283±0.012. 9.3. 4.50±0.60. 0.102±0.011. 22. β-hP-PST P236C α-hDHEA-ST I66C/K227C β-hDHEA-ST I66C/K227C. 0.64±0.04. 0.022±0.0003 0.031±0.001 0.058±0.001. 34 37 87. 4.86±0.99 8.04±1.22 14.1±4.91. 0.005±0.0003 0.032±0.001 0.029±0.004. 1.0 4.0 2.1. 0.89±0.10 1.05±0.18 0.93±0.07. 0.018±0.0005 0.028±0.001 0.038±0.0005. 20 27 41. 0.84±0.15 0.67±0.07. a. Enzyme activities were determined as described under experimental procedures. The mean ± standard error was obtained from nonlinear regression program.. b. The STs were passed through a HiTrap™ desalting column to remove small molecules and to exchange buffer (100 mM bis-tris propane at pH 7.0, 1 mM EDTA,. 10% glycerol, and 125 mM sucrose). Desalted enzymes were incubated with 1 mM TCEP at 25℃ for six hours, then passed through HiTrap™ desalting column to remove small molecules. c. The desalted β-rPST wild type and human ST mutants were incubated with 1mM GSSG for one and six hours at 25℃, respectively. Then samples were passed. through HiTrap™ desalting column. d. The oxidized STs were incubated with 1mM TCEP for six hours at 25℃, then passing through HiTrap™ desalting column.. 33.
(43) Figure 1: Physiological catalyzed by STs. PAPS is used by ST as a donor of sulfuryl group to a specific substrate. Standard assay of ST was performed using pNP as sulfuryl group acceptor described in Experimental Procedures.. 34.
(44) Figure 2: Assay of STs. The scheme for the assay of enzymatic activity. (A) The transfer reaction was catalyzed by PST that illustrated in experimental procedure. (B) Coupled-enzyme assay for DHEA-ST (or AST). The activity of hDHEA-ST for physiological reaction is to be determined shown in this figure, rPST mutant K65E/R68G represented an auxiliary enzyme used to generate PAPS. The product, MU, was used as a fluorescent indicator of enzyme turnover. This assay was based on the regeneration of PAPS from PAP catalyzed by recombinant rPST mutant using MUS as the sulfate group donor. 35.
(45) Figure 3: Multiple sequences alignment of wild type STs hSULT1A1 hSULT1A3 rPST rSULT1E1 mSULT1E1 hSULT1E1 hSULT2A1 hSULT2A3 rSULT2A1. MELIQDTSRPPLEYVKGVPLIKYFAEALGPLQSFQARPDDLLISTYPKSGTTWVSQILDMIYQGGDLEKCHRAPIFMRVPFLEF MELIQDTSRPPLEYVKGVPLIKYFAEALGPLQSFQARPDDLLINTYPKSGTTWVSQILDMIYQGGDLEKCNRAPIYVRVPFLEV ----MEFSRPPLVHVKGIPLIKYFAETIGPLQNFTAWPDDLLISTYPKSGTTWMSEILDMIYQGGKLEKCGRAPIYARVPFLEF METSMPEYYEVFGDFHGVLMDKLFTKYWEDVETFSARPDDLLVVTYPKSGSTWIGEIVDMIYKEGDVEKCKEDAIFNRIPYLEC METSMPEYYEVFGEFRGVLMDKRFTKYWEDVEMFLARPDDLVIATYPKSGTTWISEVVYMIYKEGDVEKCKEDAIFNRIPYLEC -MNSELDYYEKFEEVHGILMYKDFVKYWDNVEAFQARPDDLVIATYPKSGTTWVSEIVYMIYKEGDVEKCKEDVIFNRIPFLEC ----MSDDFLWFEGIAFPTMGFRSETLRKVRDEFVIRDEDVIILTYPKSGTNWLAEILCLMHSKGDAKWIQSVPIWERSPWVES ----MSDDFLWFEGIAFPTMGFRSETLRKVRDEFVIRDEDVIILTYPKSGTNWLAEILCLMHSKGDAKWIQSVPIWERSPWVES ----MMSDYNWFEGIPFPAISYQREILEDIRNKFVVKEEDLLILTYPKSGTNWLNEIVCLIQTKGDPKWIQTVPIWDRSPWIET. hSULT1A1 hSULT1A3 rPST rSULT1E1 mSULT1E1 hSULT1E1 hSULT2A1 hSULT2A3 rSULT2A1. KAPGIPSGMETLKDTPAPRLLKTHLPLALLPQTLLDQKVKVVYVARNAKDVAVSYYHFYHMAKVHPEPGTWDSFLEKFMVGEVS NDPGEPSGLETLKDTPPPRLIKSHLPLALLPQTLLDQKVKVVYVARNPKDVAVSYYHFHRMEKAHPEPGTWDSFLEKFMAGEVS KCPGVPSGLETLEETPAPRLLKTHLPLSLLPQSLLDQKVKVIYIARNAKDVVVSYYNFYNMAKLHPDPGTWDSFLENFMDGEVS RNEDLINGIKQLKEKESPRIVKTHLPAKLLPASFWEKNCKIIYLCRNAKDVVVSYYYFFLIIKSYPNPKSFSEFVEKFMEGQVP RNEDLINGIKQLKEKESPRIVKTHLPPKVLPASFWEKNCKMIYLCRNAKDVAVSYYYFLLMITSYPNPKSFSEFVEKFMQGQVP RKENLMNGVKQLDEMNSPRIVKTHLPPELLPASFWEKDCKIIYLCRNAKDVAVSFYYFFLMVAGHPNPGSFPEFVEKFMQGQVP -----EIGYTALSETESPRLFSSHLPIQLFPKSFFSSKAKVIYLMRNPRDVLVSGYFFWKNMKFIKKPKSWEEYFEWFCQGTVL -----EIGYTALSESESPRLFSSHLPIQLFPKSFFSSKAKVIYLMRNPRDVLVSGYFFWKNMKFIKKPKSWEEYFEWFCQGTVL -----EIGYPAIINKEGPRLITSHLPIHLFSKSFFSSKAKAIYLMRNPRDILVSGYFFWGNTNLVKNPGSLGTYFEWFLQGNVL. hSULT1A1 hSULT1A3 rPST rSULT1E1 mSULT1E1 hSULT1E1 hSULT2A1 hSULT2A3 rSULT2A1. YGSWYQHVQEWWELSRTHPVLYLFYEDMKENPKREIQKILEFVGRSLPEETVDFVVQHTSFKEMKKNPMTNYTTVPQEFMDHSI YGSWYQHVQEWWELSRTHPVLYLFYEDMKENPKREIQKILEFVGRSLPEETMDFMVQHTSFKEMKKNPMTNYTTVPQELMDHSI YGSWYQHVKEWWELRHTHPVLYLFYEDIKENPKREIKKILEFLGRSLPEETVDSIVHHTSFKKMKENCMTNYTTIPTEIMDHNV YGSWYDHVKSWWEKSKNSRVLFMFYEDMKEDIRREVVKLIEFLERDPLAELVDKIIQHTSFQEMKNNPCTNYSMLPETMIDLKV YGSWYDHVKAWWEKSKNSRVLFMFYEDMKEDIRREVVKLIEFLERKPSAELVDRIIQHTSFQEMKNNPSTNYTMMPEEMMNQKV YGSWYKHVKSWWEKGKSPRVLFLFYEDLKEDIRKEVIKLIHFLERKPSEELVDRIIHHTSFQEMKNNPSTNYTTLPDEIMNQKL YGSWFDHIHGWMPMREEKNFLLLSYEELKQDTGRTIEKICQFLGKTLEPEELNLILKNSSFQSMKENKMSNYSLLSVDYVVDKA YGSWFDHIHGWMPMREEKNFLLLSYEELKQDTGRTIEKICQFLGKTLEPEELNLILKNSSFQSMKENKMSNYSLLSVDYVVDKA FGSWFEHVRGWLSMREWDNFLVLYYEDMKKDTKGTIKKICDFLGKNLGPDELDLVLKYSSFQAMKENNMSNYSLIKEDRVTNGL. hSULT1A1 hSULT1A3 rPST rSULT1E1 mSULT1E1 hSULT1E1 hSULT2A1 hSULT2A3 rSULT2A1. SPFMRKGMAGDWKTTFTVAQNERFDADYAEKMAGCSLSFRSEL SPFMRKGMAGDWKTTFTVAQNERFDADYAEKMAGCSLSFRSEL SPFMRKGTTGDWKNTFTVAQNERFDAHYAKTMTDCDFKFRCEL SPFMRKGIVGDWRNHFPEALRERFEEHYQRHMKDCPVKFRAEL SPFMRKGIIGDWKNHFPEALRERFDEHYKQQMKDCTVKFRMEL SPFMRKGITGDWKNHFTVALNEKFDKHYEQQMKESTLKFRTEI Q-LLRKGVSGDWKNHFTVAQAEDFDKLFQEKMADLPRELFPWE Q-LLRKGVSGDWKNHFTVAQAEDFDKLFQEKMADLPRELFPWE K-LMRKGTTGDWKNHFTVAQAEAFDKVFQEKMAGFPPGMFPWE. There were nine cytosolic sulfotransferase protein sequences in National Center for Biotechnology Information (NCBI). According to multiple sequence alignment by the CLUSTALW v1.8 program at Protein Information Resource (PIR), Cys66 and Cys232 of rPST corresponding to the conserved residues of other sequences were showed in the shadows. 36.
(46) Figure 4: Molecular homologous modeling structure of rPST by SWISS-MODEL. Molecular homologous modeling mimicked rPST structure using hP-PST wild type (PDB code: 1LS6) as template by SWISS-MODEL. 37.
(47) Figure 5: Structural comparison of rPST and hSTs by Combinatorial Extension (CE) method. Panel A: Molecular homologous modeling rPST structure compared with hP-PST wild type (PDB code: 1LS6). The red, yellow, green, and blue residues represented Cys66 and Cys232 of rPST, and Cys70 and Pro236 of hP-PST, respectively. Panel B: There was structural alignment of rPST and hDHEA-ST wild type (PDB code: 1EFH). The red, yellow, green, and blue residues represented Cys66 and Cys232 of rPST, and Ile66 and Lys227 of hDHEA-ST, respectively. 38.
(48) Figure 6: The distance between relative two thiol groups of rPST and hST models. Panel A and B: Molecular homologous modeling mimicked rPST wild type and hP-PST mutant P236C structures using hP-PST wild type (PDB code: 1LS6) as template by SWISS-MODEL, and the distances between two thiol groups of the relative cysteines on rPST and hP-PST were both 12.82 Å. Panel C: Molecular homologous modeling mimicked hDHEA-ST mutant I66C/K227C structure using hDHEA-ST wild type (PDB code: 1EFH) as template by SWISS-MODEL, and the distance between two thiol groups of Cys66 and Cys227 was 11.82 Å. 39.
(49) Figure 7: Protein electrophoresis analysis of all sulfotransferase after redox treatment by SDS-PAGE. There were the SDS-gel patterns of reduced and oxidized enzymes which were treated as illustration in experimental procedure. Line 1, 3, 5, 7, 9, 12, 14, 16, 18 represented reduced forms of rPST wild type, C66S, C232S, hP-PST wild type, P236C, hDHEA-ST wild type, I66C, K227C, and I66C/K227C, respectively. The other lines represented oxidized forms of all sulfotransferases except line 11 was protein marker which molecular weight was 97, 66, 45, 30, 20, and 14 kDa, respectively.. 40.
(50) Appendix A. Equipments Equipment. Company. 550 Sonic Dismenbrator. Fisher Scientific. Fast Protein Liquid Chromatography (FPLC). Pharmacia (Hong Kong). Column. Amicon or Pharmacia. GSTrap™ Fast Flow. Pharmacia (Hong Kong). HiTrap™ desalting column. Pharmacia (Hong Kong). Commercial Blendor Incubator. Waring. Orbital Shaker Incubator. DEHG YNG Instruments Co., LTD.. DNA mini-electrophoresis tank. MUPID-2. Biologic low pressure liquid chromatography system. Bio-Rad Co., LTD.. Instant camera. Polaroid. U-3300 UV/VIS Spectrophotometer. Hitachi. F-4500 Fluorescence Spectrophotometer. Hitachi. Orbital Shaker Incubator. DEHG YNG. Ultrafiltration System. Amicon. Eppendrof centrifuge 5410. Eppendorf. High-speed centrifuge (himac CR 22G). Hitachi. Laminer Flow. Hai-Tain Scientific CO., LTD.. PCR Mechaine (Perkin Elmer 2400). Perkin Elmer. pH meter. Fisher Scientific. Water bath incubator. MEDCLUB Scientific CO., LTD.. Dry bath incubator. Violet Bio Sciences Inc.. Protein electrophoresis tank. Bio-Rad Co., LTD.. 41.
(51) B. Bacteria and plasmids a. Escherichia coli DH5α and BL21 (DE3) are used for production of a large number of plasmids and expression of proteins, respectively. Purchase them in Research Innovation Development of Food Industry Research and Development Institute. b. The wild type and mutant strains of rat phenol sulfotransferases (rPST) are gifted by American NIH Drs. W. B. Jakoby and D. Marshall. c. The clones of hP-PST (SULT1A1) and hDHEA-ST (SULT2A1) wild type are gifted by American Dr. Ming-Chih Liu.. C. Mini-preparation of E. coli plasmid DNA Use VIOGENE DNA/RNA Extraction Kit to collect the plasmid of E. coli. in terms of the alkaline lysis method of Sambrook et al. 1. Grow 5 ml plasmid-containing bacterial cells in LB (Lucia-Bertani) liquid medium (1% tryptone, 0.5% yeast extract, 0.5% NaCl) with appropriate antibiotic (ampicillin, 250 μg/ml) overnight (12-16 hours) with vigorous agitation (150 rpm) at 37°C. 2. Pellet the cells by centrifuging at 12,000 × g for one minute. Decant the supernatant and remove all medium residues by pippet. 3. Add 250 μl of MX1 Buffer to the pellet, and re-suspend the cells completely by vortexing or pipetting. 4. Add 250 μl of MX2 Buffer and gently mix (invert the tube 4-6 times) to lyse the cells until the lysate becomes clear. Incubate at room temperature for 1-5 minutes. 5. Add 350 μl of MX3 Buffer to neutralize the lysate, then immediately and gently mix the solution. A white precipitate should be formed. 42.
(52) 6. Centrifuge for ten minutes, meanwhile place a Mini-M™ Column onto a collection tube. Then transfer the supernatant carefully into the column. 7. Centrifuge for one minute. Discard the flow-through. 8. Wash the column once with 0.5 ml WF Buffer by centrifuging for one minute. Discard the flow-through. 9. Wash the column once with 0.7 ml WS Buffer by centrifuging for one minute. Discard the flow-through. And Centrifuge the column at full speed (12,000 × g) for another three minutes to remove residual ethanol. 10. Place the column onto a new 1.5-ml centrifuge tube. Add 50 μl of Elution Buffer onto the center of the membrane. 11. Stand the column for 1-2 minutes, and centrifuge for one minute to elute DNA. And store plasmid DNA at 4°C or -20°C. D. Mutant strand synthesis reaction 1. Prepare the ds-DNA template either by standard miniprep protocols or by cesium chloride gradient purification. 2. Prepare the mutant strand synthesis reactions for thermal cycling as indicated in the table below, according to the template size and number of mutagenic primers. Add the components in the order listed then mix gently by pipetting or tapping the reaction tube. Parameter Extension time PfuTurbo® DNA polymerase Input template Primers (each) dNTP concentration Denaturing temperature Extension temperature. <10 kb plasmid DNA. >10 kb plasmid DNA. 1 minute per kb 2.5 U 50-100 ng plasmid DNA 100-200 ng (0.2-0.5 μM) 100-250 μM each dNTP 95°C 72°C. 2 minutes per kb 5.0 U 200-250 ng plasmid DNA 200 ng (0.5 μM) 500 μM each dNTP 92°C 68°C. Total reaction volume: 50 μl 43.
(53) 3. Cycle the reactions using the cycling parameters outlined in the table below. <10 kb plasmid DNA Segment. Number of cycles. Temperature. Duration. 1. 1. 95°C. 2 minutes. 95°C. 30 seconds. Primer Tm-5°C. 30 seconds. 72°C. 1 minute for targets ≤1 kb 1 minute per kb for targets >1 kb. 1. 72°C. 10 minutes. Segment. Number of cycles. Temperature. Duration. 1. 1. 92°C. 2 minutes. 92°C. 10 seconds. Primer Tm-5°C. 30 seconds. 68°C. 2 minutes per kb. 92°C. 10 seconds. Primer Tm-5°C. 30 seconds. 68°C. 2 minutes per kb plus 10 seconds/cycle. 2. 30. 3 >10 kb plasmid DNA. 2. 10. 3. 20. 4. Following temperature cycling, place the reaction on ice for 2 minutes to cool the reaction to ≤37°C.. E. Dpn I digestion of the amplification products 1. Add 1 μl of Dpn I restriction enzyme (10 U/μl) directly to each amplification reaction tube. 2. Gently and thoroughly mix each reaction mixture by pipetting the solution up and down several times. Spin down the reaction mixtures in a micro-centrifuge for one minute, then immediately incubate each reaction at 37°C for 12 hours to digest the parental (nonmutated) ds-DNA.. 44.
(54) F. Transformation 1. Gently thaw the CaCl2-treated competent cells which stored at -80°C on ice. 2. Transfer 5 μl of the Dpn I-treated DNA from each mutagenesis reaction to competent cells. 3. Swirl the transformation reactions gently to mix, and then incubate the reactions on ice for 30 minutes. 4. Heat-pulse the tubes in at 42°C water bath for 45 seconds, then put the tubes on ice immediately for 2 minutes. 5. Add 0.5 ml of LB medium to each tube and incubate the tubes at 37°C for one hour with shaking at 150 rpm. 6. Plate the appropriate volume (100 μl) of each transformation reaction on agar plates containing the appropriate antibiotic (250 μg/ml) for the plasmid vector. 7. Spread cells on LB-ampicillin agar plates and incubate at 37°C overnight.. G. Protein expression and purification a. Recombinant rPST 1. Inoculate 5 ml of LB-medium containing 250 μg/ml ampicillin in 15-ml class tube with a colony of E. coli. containing the constructed recombinant expression plasmid (pET-3c/rPST). Incubate the cultures overnight at the appropriate temperature (37°C). 2. Inoculate 50 ml of LB-medium containing 250 μg/ml ampicillin in 100-ml flask with 1 ml of overnight culture of E. coli. Incubate the cultures at 37°C in a shaking incubator (150 rpm) until cells reach mid-log phase of growth (A600 ≈ 0.5).. 45.
數據

+7
相關文件
中興國中
中興國中
使用限制 海水、硬水 皂化反應.
溶液中離子沈澱反應的化學反應式表示法:以氯化鈉和硝酸銀的反應為例(如上圖)
酸性氣體(二氧化硫、二氧化氮)可以飄浮到離源頭很遠的地
而在後續甲烷化反應試驗方面,以前段經厭氧醱酵產氫後之出流水為進流基 質。在厭氧光合產氫微生物方面,以光合作用產氫細菌中產氫能力最好的菌株 Rhodopseudomonas palustris
SF15140A 楊勝舜 利用 JKB-122 評估對於經干擾素(長效 型或短效型)或干擾素和 Ribavirin 組合 治療沒有反應的 C 型肝炎病毒陽性患 者之肝臟功能 (丙胺酸轉胺酶
化學風化作用(Chemical Weathering) :係岩石被溶解、氧化及