行政院國家科學委員會專題研究計畫 期中進度報告
斜紋夜蛾微粒子蟲之研究: 核醣體, 極管蛋白及孢壁蛋白
基因(2/3)
計畫類別: 個別型計畫
計畫編號: NSC92-2313-B-002-052-
執行期間: 92 年 08 月 01 日至 93 年 07 月 31 日
執行單位: 國立臺灣大學昆蟲學系暨研究所
計畫主持人: 王重雄
報告類型: 精簡報告
處理方式: 本計畫可公開查詢
中 華 民 國 93 年 5 月 31 日
行政院國家科學委員會補助專題研究計畫
□ 成 果 報 告
■期中進度報告
斜紋夜蛾微粒子蟲: 核醣體,極管蛋白基因及孢壁蛋白基因
計畫類別:■ 個別型計畫 □ 整合型計畫
計畫編號:NSC 92-2313-B-002-030-
執行期間: 92 年 08 月 01 日至 93 年 07 月 31 日
計畫主持人:王重雄
共同主持人:
計畫參與人員:
成果報告類型(依經費核定清單規定繳交):■精簡報告 □完整報告
本成果報告包括以下應繳交之附件:
□赴國外出差或研習心得報告一份
□赴大陸地區出差或研習心得報告一份
□出席國際學術會議心得報告及發表之論文各一份
□國際合作研究計畫國外研究報告書一份
處理方式:除產學合作研究計畫、提升產業技術及人才培育研究計畫、
列管計畫及下列情形者外,得立即公開查詢
□涉及專利或其他智慧財產權,□一年□二年後可公開查詢
執行單位:台灣大學昆蟲系暨研究所
中 華 民 國 九十三 年 五 月 三十一 日
中文摘要
斜紋夜蛾微粒子蟲及微粒子蟲屬模式種家蠶微粒子蟲 (Nosema bombycis),其完整核醣體
RNA 基因序列 (ribosomal RNA genes, 分別為 4,296 及 4,301 bp) 完成全長定序,家蠶微粒
子蟲(N. bombycis)核醣體基因已完成分析其結構,經分析其基因序列自 5’端起包含
LSUrRNA (large subunit ribosomal RNA, 2,497 bp), ITS (inter-transcribe space, 179bp),
SSUrRNA (small subunit ribosomal RNA, 1,232 bp), IGS (intergenic spacer, 279) 以及 5S
rRNA (114 bp)。本文內亦構築及討論家蠶微粒子蟲 LSUrRNA 之二級結構。其核醣體基因
的排列為 LSUrRNA – ITS – SSUrRNA – IGS – 5S,這種排列方式為家蠶微粒子蟲所特有,
並未在其他微孢子蟲中被發現。已知的微孢子蟲核醣體基因排列方式皆為 SSU-ITS-LSU,
其中還包括微粒子蟲屬中的 Nosema apis,在模式種 (N. bombycis) 中所發現之特有 rRNA
基因排列方式,或許在微粒子蟲屬分類上有其重要性。部分結果已發表於 Fungal Genetic and
Biology 41 卷,473 至 481 頁。
Abstract
The complete DNA sequences data of the ribosomal RNA (rRNA) genes of the microsporidian type
species,
Nosema spodopterae
, and
N. bombycis
. Sequences for the
N. bombycis
rRNA large subunit
gene (LSUrRNA: 2497 bp, GenBank Accession No.
AY211393
), the internal transcribed spacer (ITS:
179 bp, GenBank Accession No.
AY211394
), the small subunit gene (SSUrRNA: 1232 bp), intergenic
spacer (IGS: 279 bp), and 5S region (114 bp) are also given, and the secondary structure of the large
subunit is discussed. The unusual organization of the
N. bombycis
rRNA genes, LSUrRNA- ITS-
SSUrRNA- IGS -5S, is the first found in microsporidia. This novel arrangement, in which the LSU is
located at 5
'end of the SSU, is the reverse of the organizational sequence (i.e., SSU-ITS-LSU) found
in all previously reported microsporidian rRNAs, including
Nosema apis
. This unique character in the
type species may have taxonomic implications for the members of the genus
Nosema
. This result has
been published in the journal, Fungal Genetic and Biology 41, 473-481 pp.
Introduction
Microsporidia are intimate parasites of animals, well adapted in pathogenicity transmission, ecology, and resistance to the immunity of their host. They are small unicellular protists and obligate intracellular parasites. Insects in nearly all taxonomic orders are susceptible to this pathogen, but over half of the susceptible insect hosts occur in two orders, Lepidoptera and Diptera. Most of the entomopathogenic microsporidia occur in the genus Nosema. N. bombycis is a type species of genus Nosema (Sparague et al., 1992). This microsporidium has caused a heavy loss of sericulture in Europe, especially at as well as in Asia and America, especially in the meddle of 19th century (Steinhaus, 1963).
Microsporidia are extremely ancient eukaryotes and unusual in lacking mitochondria. Accumulating molecular data and phylogenic analyses by computer seem to suggest that mitochondrial endosymbiosis occurred before the emergence of microsporidia (Germot et al., 1997) and also microsporidia share a common origin with fungi. They are, therefore, most probably just a curious type of fungi (Van de Peer, 2000). They present prokaryote-like features in their rRNA gene organization and sequence (Vossbrink et al., 1987; Galtier & Gouy, 1995). No distinct 5.86 rRNA gene has been found in the reported microsporidian rRNA gene (Vossbrinck & Woese, 1986; Gatehouse & Malone, 1998), and the sequences of microsporidian rRNA are shorter than the known sequences of eukaryotic or prokaryotic rRNA (De Rijk et al., 1998; Tsai et al., 2002). In contrast to the SSUrRNA sequences of microsporidia in GenBank, only four complete LSUrRNA gene sequences of microsporidia─ Encephalitozoon cuniculi (Peyretaillade et al., 1998), Microsporidium 57864 (GenBank Accession No. U90885), N. apis (Gatehouse & Malone, 1998), and Heterosporis anguillarum (Tsai et al., 2002) ─are published and registered in GenBank. With such limited available data of the complete sequences of microsporidian rRNA, it implies that microsporidia have a high diversity in LSUrRNA sequence and also rRNA gene organization.
N. bombycis was the first known microsporidium discovered in the early 19th century when an
epidemic microsporidiosis ravaged the silkworm industry of Europe. This microsporidium was named and classified as a type species of genus Nosema by Nägeli (1857) (Sparague, 1992). Much research concerning aspects of this microspodium have been published, but only uncompleted sequences of N.
bombycis rRNA genes were found in the literature, SSUrRNA sequence (1,232 bp) and a partial
LSUrRNA (292 bp) of N. bombycis are found in GenBank (Accession No. D85503 and L28962) (Hatakeyama et al., 1997; Baker et al., 1994). As part of an effort to carry out the complete sequence of N.
bombycis rRNA gene for the study of the member of genus Nosema, we present the complete rDNA
sequence and gene organization of N. bombycis rRNA genes, including LSUrRNA gene, internal transcribed spacer (ITS), SSUrRNA gene, IGS I and 5S gene. In addition to the sequences and organization, the secondary structures of the N. bombycis rRNA genes are also constructed and discussed.
Materials and methods
Spore purification and nucleic acid preparation.
Microsporidian spores of N. bombycis were a gift from Dr. R. Sugimoto the MAFF GENE Bank of the National Institute of Agrobiological Science, Japan. The purification of spores was carried out as described previously (Huang et al., 1998; Tsai et al., 2002).
Amplification and sequencing strategy of rRNA genes
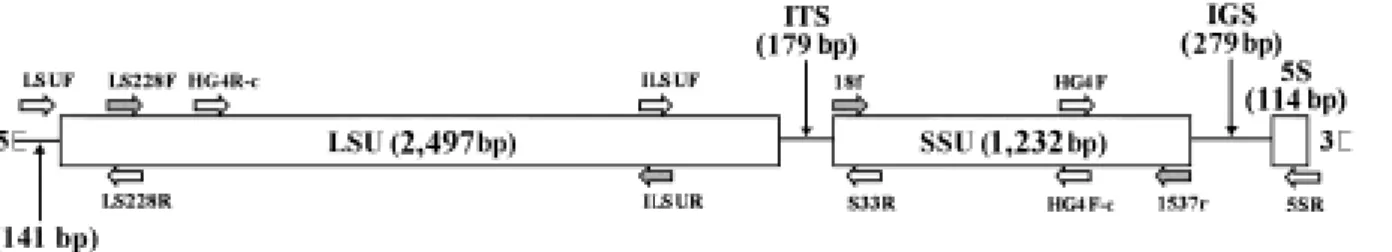
The primer sets used for rRNA gene amplification and the amplicon’s sizes with primer sets are shown in Table 1 and Fig. 1. The coding region of SSU rDNA of N. bombycis was amplified by using a primer set: 18f and 1537r (Vossbrinck et al., 1993). The major coding region of LSU rDNA was amplified by a primer set: LS228F and ILSUR (Vossbrinck et al., 1993). The 5’ end of LSU rDNA was achieved by a primer set: LSUF and HG4R (Gatehouse & Malone, 1998). The 3’ end of LSU rDNA and ITS (internal transcribed spacer) were amplified by a primer set: ILSUF and S33R. The ILSUF primer is the complement sequence of ILSUR (Tsai et al., 2002). The 5S and IGS were amplified by a primer set: HG4F (Gatehouse & Malone, 1998) and 5SR. The primer 5SR was designed by the conserved region of 5S.
PCR amplification, cloning and sequencing of rRNA gene
The genomic DNA (80 ng) of N. bombycis was mixed in a 100 µl PCR reaction mixture containing 10 mM Tris-HCl, pH 9.0, 50 mM KCl, 1.5 mM MgCl2, 100 mM of each dNTP, 100 pmol of each primer (Table 1), and 2.5 units Taq DNA polymerase (Promega). The amplification was performed in an AG-9600 Thermal Station (Biotronics Corp.) for 40 cycles, each with the following profile: 94 ˚C for 0.5
min, 50 ˚C for 0.5 min, and 72 ˚C for 2 min. A 10 µl aliquot from each reaction was run on a 1.0% agarose gel to visualize the PCR products. The gel was photographed using the Eagle-Eye II photo-documentation system (Stratagene). The PCR products were eluted by an E.Z.N.A. Gel Extraction Kit (Omega Bio-tek). The eluted DNAs were then sequenced directly on an automated DNA Sequencer (DNA Sequencer 377, Applied Biosystems).
Confirmation of the rRNA gene organization of N. bombycis
The total length of rRNA of N. bombycis was amplified by a primer set: LSU/5SR with Platinum Pfx
DNA polymerase (Invitrogen). The amplicon was then eluted and used as a template to amplify fragments with several primer sets, HG4R-c/HG4F-c and ILSUF/HG4F-c for partial LSUrRNA-ITS-partial
SSUrRNA; ILSUF/1537R for partial LSUrRNA-ITS-SSUrRNA; and ILSUF/5SR for partial LSUrRNA-ITS-SSUrRNA-IGS-5S. The amplification method was similar to previous description.
Secondary structure construction
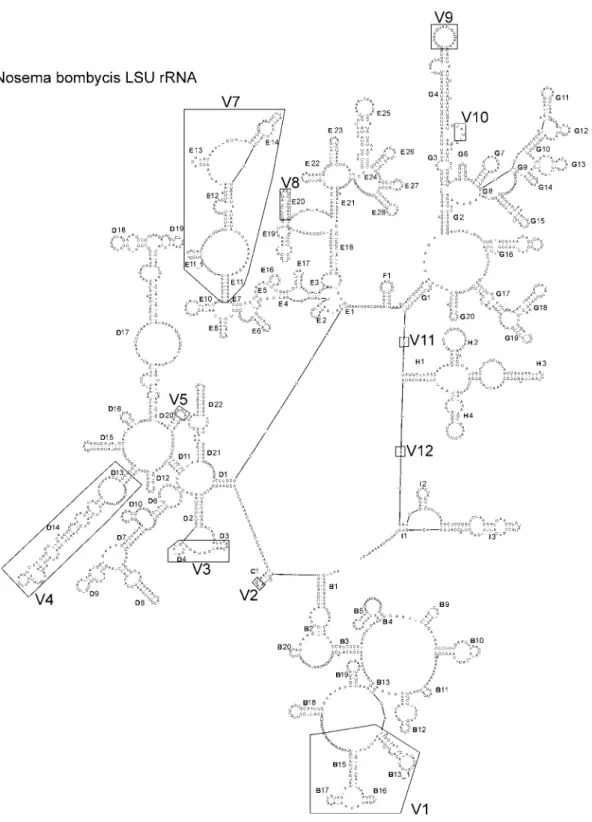
The secondary structures of N. bombycis LSUrRNA were constructed by a manual and automatic method starting from DCSE alignment files (De Rijk & De Wachter, 1993) and drawn by the RnaViz program (De Rijk & De Wachter, 1997, De Rijk & De Wachter, 2003). The secondary structures of N.
bombycis SSUrRNA can be found in the European sallsubunit ribosomal RNA database (Van de Peer et al., 2000). The N. bombycis LSUrRNA were aligned to the rRNA database. The helices in the rRNA
secondary structure elements were localized and given a number named in all known eukaryotic LSUrRNA (V1-12 in eukaryotic LSUrRNA).
Results
The complete sequence and organization of Nosema. bombycis rRNA
The complete DNA sequence of N. bombycis rRNA gene was carried out and the organization was analyzed (Fig. 1). The complete DNA sequence of N. bombycis rRNA gene contained 4,301 bp (GenBank Accession No. AY259631), and the organization of N. bombycis rRNA gene from 5’end consisted of the large subunit gene (LSUrRNA: 2,497 bp) (GenBank Accession No. AY211393), the internal transcribed spacer (ITS: 179 bp) (GenBank Accession No. AY211394), the small subunit gene (SSUrRNA: 1,232 bp), intergenic spacer (IGS: 279 bp), and 5S (114bp). The PCR results with primer sets (Table 1) are shown in Fig. 1.
LSUrRNA gene sequence
The main part of the LSUrRNA gene was amplified with a primer set (LS228F and ILSUR), and six internal sequencing primers were used for the sequencing of the amplicon, the size of the amplicon was 2,108 bp. For sequencing 5’ end of LSUrRNA, we used a primer set (LSUF and HG4R). The putative start and terminal regions were determined by comparison to N. apis LSUrRNA sequence and the secondary structure construction of N. bombycis LSUrRNA gene (Fig 2). The LSUrRNA gene contains 2,497 bp, and the base composition of the LSUrRNA sequence is 31.9% G + C. It is the lowest G + C content of all known microsporidian LSUrRNA genes. The sequence identities of LSUrRNA sequences between N. bombycis and N. apis (GenBank Accession No. U97150) (Gatehouse & Malone, 1998), or Microsporidium 57864 (GenBank Accession No. U90885), H. anguillarum (GenBank Accession No. AF402839) (Tsai et al., 2002), and Encephalitozoon cuniculi (GenBank Accession No. AJ005581) (Peyretaillade et al., 1998) were 71%, 69%, 46%, and 53%, respectively. This sequence was compared with the reported incomplete sequences of LSUrRNA genes, especially with the partial sequence of N.
bombycis LSUrRNA gene reported by Baker et al. (1994) (GenBank Accessory No. L28962). Those
sequences are located at 132 to 423bp from the 5’ end of our sequence (100% identity), and their 580R primer is partially identified with HG4R primer. The DNA sequence of LSUrRNA gene of N. bombycis has been submitted to GenBank with an accessory number is AY211393.
The internal spacer of N. bombyc lacks the 5.8S rRNA gene. The sequence of the LSUrRNA gene, located at 1 to 160 nucleotide from the 5’end, corresponds to the known fungal 5.8S rRNA sequences,
Cystofilobasidium bisporidii (Accessory No. M94511), Lactarius acerrimus (AJ278139), Thanatephorus cucumeris (AB019008), Trichoderma reesei (L27800), and Tuber cf. rapedorum (AJ278140). The
homologies are 34%, 44%, 44%, 44%, and 42%, respectively by Clustal X and GeneDoc.
The secondary structure of the LSUrRNA of N. bombycis (Fig. 2) is basically similar to that of N.
apis and H. anguillarum (Tsai et al., 2002; De Rijk et al., 1998). Based on the secondary structures of the
eukaryotic LSUrRNA of Xenopus laevis (De Rijk & De Wachter, 1997), seven groups (B to I) can be distinguished clockwise from a core area. Fifteen hypervariable areas (V1-12) are also shown in Fig. 2.
Nine helices (B6, B7, B8, B14, B21, D5, E9, E15, and G5) are missing. Four areas of the hypervariable areas are also almost entirely missing (V2, V8, V10 and V12), and four areas are extremely reduced (V1, V3, V5 and V6). Comparing the secondary structure of N. bombycis LSUrRNA with those of other known species, N. apis, Microsporidium 57864, E. cuniculi, and H. anguillarum, the most divergence occurs in the V4 area. N. bombycis and E. cuniculi LSUrRNAs contain specific conformations in the V3 area while four added areas (V5-9) in H. anguillarum LSUrRNA show their specific conformations.
ITS sequence
In contrast to all the known rRNA genes, the ITS region of N. bombycis is localized between LSU and SSUrRNA. It contains 179 bp located 2498 to 2576 nucleotides from the 5’ end of the rRNA gene (Fig. 1). The G + C content of the ITS sequence is 19.6%. The DNA sequence has been submitted to GenBank, with the accessory number AY211394.
SSUrRNA gene sequence
The SSUrRNA gene contains 1232 bp located 2677 to 3908 nucleotides from the 5’ end of the rRNA gene (Fig.1). The G + C content of SSUrRNA gene is 34.2%. The complete DNA sequence of the SSUrRNA gene of N. bombycis had a 99% homology to that of N. bombycis in GenBank (Accessory No. D85503) but also to the other isolate (D85504) (Hatakeyama et al., 1997). Only two nucleotides are different from D85503, located at 3497 and 3874.
IGS
A 279 bp sequence located between SSU and 5S rRNA genes, this fragment was named the IGS region. It contains 30% G + C and is located at nucleotides 3909 to 4187. Homology is not high (only 20 nucleotides overlapping) with other known microsporidian ITS or IGS sequences by standard
nucleotide-nucleotide BLAST [blastn], Nucleotide BLAST, NCBI.
5S rRNA gene
The 5S rRNA of N. bombycis complete sequence has 114 bp (with putative end), located at 4188 to 4301 nucleotides from the 5’ end of the N. bombycis rRNA. The G + C content of 5S rRNA is 47.3%. The sequence was compared with two other N. bombycis 5S rRNAs (Accession No. D14631 and AB097401) (Kawakami et al., 1992) and showed homologies of 91% (only 10 nucleotides difference) and 92% (only 9 nucleotides difference), respectively, but compared with Microsporidium 57864 (Accession No. U90885) homology was only 77%.
Confirmation of the rRNA gene organization of N. bombycis
To confirm the organization of N. bombycis rRNA gene as shown in Fig. 1, the whole rRNA gene was amplified with LSUF/ 5SR primer set; the amplicon size was matched to the sequenced length (4,442 bp) (Fig. 3A), and the other four primer sets were used to amplify the internal fragments of rRNA. The amplicon with a primer set HG4F-c/ HG4R-c, complementary sequences of HG4F and HG4R, is 3,031 bp (Fig. 3B, lane 1), and the other three primer sets, ILSUF/HG4F-c, ILSUF /1537r, and ILSUF/5SR yield 1,261 bp, 1,700 bp, and 2,093 bp, respectively (Fig. 3 B, lane 2-4). These PCR products are matched to the size of the sequenced data. These products were sequenced, and the results were similar to previous data. In addition, we tried to amplify the supposed IGS region between SSU and LSU by PCR, but no product was found.
Discussion
Microsporidia are tiny eukaryotic organisms and infect all major animal groups. The phylum Microsporia contains 143 genera, more than 1200 species. The small genomic size (2.9-19.5 Mb) of these organisms indicates that they may have developed strategies of packing genetic information tightly into the genome or they may have lost genetic information for a metabolic pathway and depend on host cell sources for these compounds (Weiss & Vosdbrinck, 1999). The genomic study of microsporidia is currently a highly attractive topic. In fact, molecular knowledge of the microsporidia is still rather limited, but several findings evoked an evolutional concept of microsporidia, i.e., a mitochondrial-type hsp 70 found in microsporidia implies that they are derived from a mitochondriate eukaryote and close fungal relationship (Hirt et al., 1997; Williams et al., 2002). Two separate proteins of thymidylate synthase and dihydrofolic acid reductase and the sequence of tubulin genes of microsporidia confirmed a close fungal relationship (Edlined et al., 1996). Otherwise, the high variation of microsporidian genomic size leads us to believe a high divergence of gene structure and organization may occur among the member of microsporidia, during evolution.
spacer regions (ITS and IGS) around them are highly variable, even between closely related species. These highly variable and conserved rRNA gene regions provide a means for analyzing phylogenetic relationships over a wide range of taxonomic levels. The rRNA components of eukaryotes and prokaryotes differ in gene organization and size. Microsporidia present prokaryote-like features in their rRNA gene organization and sequence (Vossbrinck et al., 1987; Galtier & Gouy, 1995), but the sequences of microsporidian rRNAs are shorter than the known sequences of eukaryotic or prokaryotic rRNA (De Rijk et al., 1998). No distinct 5.8S rRNA gene is found in any reported microsporidian rRNA gene, but the homology sequences were found at the 5’end of LSUrRNA (Vossbrinck & Woese, 1986; Gatehouse & Malone, 1998). These findings imply that microsporidia contain a special rRNA gene not found in other organisms and the variation in microspridian rRNA gene may even occur.
The SSUrRNA gene sequences of microsporidian rRNA are highly conserved, and therefore sequence data for the SSUrRNA gene for a number of species is available, and it is often chosen to investigate the taxonomic position and phylogeny of microsporidia species (Vossbrinck et al., 1987; Vossbrinck et al., 1993; Visvesvara et al., 1994; Baker et al., 1995; Malone & McIvor, 1996; Pieniazek et al., 1996; Nilson
et al., 1998; Gresoviac et al., 2000). Despite this, the results give rise to several questions about whether
the taxonomic positions of several species are ambiguous (Tsai et al., 2002; Hirt et al., 1997) and whether SSUrRNA can be accurately used as an available marker for distinguishing microsporidian species. In fact, the sequence of SSUrRNA is useless in distinguishing between very closely related species, even those that can be distinguished on morphological criteria (Canning et al. 1999). Indeed, our previous papers showed that the similarity of the SSUrRNA framework in each group of microsporidia is very high, and it can work on determination of an unknown species in a high taxon and yet not work on identification of a closely related species (Tsai et al., 2002; Tsai, 2003, in press).
Only limited data of the 3’-end of SSUrRNA-ITS and partial 5’-end of the LSUrRNA genes has been published (Baker et al., 1994; Hirt et al., 1997), especially the complete sequence of LSUrRNA gene and also 5S rRNA. We have found the sequences containing internal transcribed space (ITS) and partial sequence of the 5’-end large subunit rDNA (LSUrDNA) from the Vairimorpha group (Accession No. AF141129, AF141130, AF033315, AF033316, and V. necatrix) (Vossbrinck et al., 1993) but not from the
Nosema group, except N. apis. Although a partial LSUrRNA gene sequence of N. bombycis (Accession
No. L28962) and other microsporidian species (Baker et al., 1994) has been published, the similar sequence was found in our data too (nucleotide site located from 132 to 423), but we could not find any ITS sequences of the Nosema group in the literature (except N. apis), therefore we tried to design the primer sets, based on the sequence of N. apis, for amplification of N. bombycis ITS and the 5’end or 3’end of LSUrRNA, but we failed. There is only one reason that N. apis rRNA is high divergent from N.
bombycis rRNA, except SSUrRNA gene. Indeed, N. apis has been shown to be more like some Vairimorpha species than some other Nosema species (Steinhuas, 1963; Baker et al., 1994; Kawakami et
al., 1992). Furthermore, only four microsporidian complete rRNA gene sequences have been published─
Encephalitozoon cuniculi (Peyretaillade et al., 1998), Microsporidium 57864 (GenBank Accession No.
U90885), N. apis (Gatehouse & Malone, 1998), and Heterosporis anguillarum (Tsai et al., 2002). This implies again that a vast difference in rRNA gene, especially ITS and LSUrRNA gene, exists in the course of a high reduction of their genome and gene structure during evolution. Therefore, efforts to complete the rRNA gene sequences of microsporidia will be a great contribution to understanding interspecies phylogenetic relationships. A 5.8S correspondent sequence was found from the 5’end of LSUrRNA in our datum, which is coincident with Vairimorpha necatrix (Vossbrinck & Woese, 1986; Gatehouse & Malone, 1998).
Insects in nearly all taxonomic orders are susceptible to this pathogen, and over half of the susceptible insect hosts belong to one of two orders, Lepidoptera and Diptera. Several species of microsporidia play an important role in regulating insect populations (Tanada, 1993). The members of genus Nosema are often considered the most important and widely distributed group of microsporidia. N. bombycis is a type species of the genus Nosema. Therefore, it’s characteristics are criteria for members of this genus, not only in their life cycle, development, and morphological characteristics but also in their biochemical and molecular characteristics. In this paper, we published the complete sequence of N. bombycis rRNA. Its gene organization is unique and obviously different from that of N. apis and other known species, and even from all known rRNAs. This result was confirmed again (Fig. 3A, B) and our other works on the complete sequence of N. spodopterae rRNA gene sequences (AY211390 to AY211392; unpublished data) showed the similar results. This unusual organization of N. bombycis rRNA,
LSUrRNA-ITS-SSUrRNA-IGS I-5S, will be an important characteristic of the Nosema group and also will be a great help in the Nosema species clarification through molecular markers. Furthermore, ITS and IGS share no homology with other known microsporidan sequences.
The secondary structures of N. bombycis SSUrRNA and LSUrRNA were constructed and compared their structures compared with the published data (De Rijk & De Wachter, 1997; Wuyts et al., 2001). Because the difference from the known sequence of N. bombycis SSUrRNA molecules is only two nucleotides, the whole view of secondary structure (data not shown) is similar to the structure got from the database (De Rijk et al., 1998). Several clues for criteria of microsporidian species or genera based on the LSUrRNA secondary structures of known species could be found. Due to limited available data, no solid conclusions can yet be made about the difference between LSUrRNA secondary structures within the genera or even species.
References
Baker, M.D., Vossbrinck, C.R., Maddox, J.V. and Undeen, A.H., 1994. Phylogenetic relationships among Vairimorpha and Nosema species (Microspora) based on ribosomal RNA sequence data. J. Invertebr. Pathol. 64, 100-106.
Canning, E.U., Curry,A., Cheney, S., Lafranchi-Tristem, N.J. and Haque, M.A., 1999. Vairimorpha imperfecta n.sp., a microsporidian exhibiting an abortive octosporous sporogony in Plutella xylostella L. (Lepidoptera: Yponomeutidae). Parasitology 119, 273-286.
De Rijk, P., Caers, A., Van de Peer, Y. and De Wachter, R., 1998. Database on the structure of large ribosomal subunit RNA. Nucleic Acids Res. 26, 183-186.
De Rijk, P. and De Wachter, R., 1993. DCSE, an interactive tool for sequence alignment and secondary structure research. Comput. Appl. Biosci. 9, 735-740.
De Rijk, P. and De Wachter, R., 1997. RnaViz, a program for the visualization of RNA secondary structure. Nucleic Acids Res. 25, 4679-4684.
De Rijk, P., Gatehouse, H.S. and De Wachter, R., 1998. The secondary structure of Nosema apis large subunit ribosomal RNA. Biochim. Biophys. Acta. 1442, 326-328.
De Rijk, P., Wuyts, J. and De Wachter, R. 2003. RnaViz 2: an improved representation of RNA secondary structure. Bioinformatics 19 (2), 299-300.
Edlined, T.D., Li, J., Visvesvara, G.S., Voodkin, M.H., McLaughlin, G.L. and Katiyar, S.K., 1996. Phylogenetic analysis of beta-tubulin sequences from amitochondrial protozoa. Mol. Phylognet. Evol. 5, 359-367.
Galtier, N. and Gouy, M., 1995. Inferring phylogenies from DNA sequences of unequal base compositions. Proc. Natl. Acad. Sci. USA 92, 11317-11321.
Gatehouse, H.S. and Malone, L.A., 1998. The ribosomal RNA gene region of Nosema apis (Microspora): DNA sequence for small and large subunit rRNA genes and evidence of a large tandem repeat unit size. J. Invertebr. Pathol. 71, 97-105.
Germot, A., Philippe, H. and LeGuyader, H., 1997. Evidence for loss of mitochondria from a mitochondrial-type HSP 70 kD in Nosema locustae. Mol. Biochem. Parasitol. 87, 159-168.
Gresoviac, S.J., Khattra, J.S., Nadler, S.A., Kent, M.L., Devlin, R.H., Vivares, C.P., Fuente, E.DeLa and Hedrick, R.P., 2000. Comparison of small subunit ribosomal RNA gene and internal transcribed spacer sequences among isolates of the intranuclear microsporidian Nucleospora salmonis. J. Eukaryot. Microbiol. 47, 379-387.
Hatakeyama, Y., Kawakami, Y., Iwano, H., Inoue,T. and Ishihara, R., 1997. Analyses and taxonomic inferences of small subunit ribosomal RNA sequences of five microsporidia pathogenic to the silkworm, Bombyx mori. J. Seric. Sci. Jpn. 66, 242-252.
Hirt, R.P., Healy, D., Vossbrinck, C.R., Canning, E.U. and Embley, T.M., 1997. Identification of a mitochondrial HSP 70 orthologue in Vairimorpha necatrix: molecular evidence that microsporidia once contained mitochondria. Curr. Biol. 7, 995-998.
Huang, H.W., Lo, C.F., Tseng, C.C., Peng, S.E., Chou, C.M. and Kou, C.H., 1998. The small subunit ribosomal RNA gene sequence of Pleistophora anguillarum and the use of PCR primers of diagnostic detection of the parasite. J. Eukaryot. Microbiol. 45, 556-560.
Kawakami, Y., Inoue, T., Kikuchi, M., Takayanagi, M., Sunairi, M., Ando, T. and Ishihara, R., 1992. Primary and secondary structures of 5S ribosomal RNA of Nosema bombycis (Nosematidae, Microsporidia). J. Seric. Sci. Jpn. 61, 321-329.
Malone, L.A. and McIvor, C.A., 1996. Pulsed-field gel electrophoresis of DNA from four microsporidian isolates. J. Invertebra. Pathol. 68, 231-238.
Nilsen, F., Endresen, C. and Hordvik, I., 1998. Molecular phylogeny of microsporidia with particular reference to species that infect the muscles of fish. J. Eukaryot. Microbiol. 45, 535-543.
Peyretaillade, E., Biderre, C., Peyret, P., Duffieux, F., Metenier, G., Gouy, M., Michot, B. and Vivares, C. P., 1998. Microsporidian Encephalitozoon cuniculi, a unicellular eukaryote with an unusual chromosomal dispersion of ribosomal genes and a LSUrRNA reduced to the universal core. Nucleic Acids Res. 26, 3513-3520.
trichoplusiae is a synonym of Nosema bombycis based on the sequence of the small subunit ribosomal RNA coding region. J. Invertebra. Pathol. 67, 316-317.
Steinhaus, E.A., 1963. Insect Pathology: An Advanced treatise. Academic Press, NY., AF, London.
Sprague, V., Becnel, J.J. and Hazard, E.I., 1992. Taxonomy of phylum Microspora. Crit. Rev. Microbiol. 18, 285-395.
Van de Peer, Y., De Rijk, P., Wuyts, J., Winkelmans, T. and De Wachter, R., 2000. The Europen small subunit ribosomal RNA database. Nucleic Acids Res. 28, 175-176.
Vossbrinck, C.R., Maddox, J.V., Friedman, S., Debrunner-Vossbrinck, B. A. and Woese, C.R., 1987. Ribosomal RNA sequence suggests microsporidia are extremely ancient eukaryotes. Nature 326, 411-414.
Vossbrinck, C.R. and Woese, C.R., 1986. Eukaryotic ribosomes that lack a 5.8S RNA. Nature 320, 287-288.
Tsai, S.J., Kou, G.H., Lo, C.F., and Wang, C.H., 2002. Complete sequence and structure of ribosomal RNA gene of Heterosporis anguillarum. Dis. Aquat. Org. 49, 199-206.
Tanada, Y. and Kaya, H.K., 1993. Insect Pathology. Academic Press, INC.
Tsai, S.J., Lo, C.F., Soichi, Y. and Wang, C.H., 2003. The characterization of Microsporidian isolates (Nosematidae: Nosema) from five important lepidopteran pests in Taiwan. J. Invertebr. Pathol. (in press).
Undeen, A.H. and Cockburn, A.F., 1989. The extraction of DNA from microsporidia spores. J. Invertebr. Pathol. 54, 132-133.
Visvesvara, G.S., Leitch, G.J., Da Silva, A.J., Croppo, G.P., Moura, H., Wallace, S., Slemenda, S.B., Schwartz, D.A., Moss, D., Bryan, R.T. and Pieniazek, N.J., 1994. Polyclonal and monoclonal antibody and PCR-amplified small-subunit rRNA identification of a microsporidian Encephalitozoon hellem, isolated from an AIDS patient with disseminated infection. J. Clin. Microbiol. 32, 2760-2768.
Vossbrinck, C.R., Baker, M.D., Didier, E.S., Debrunner-Vossbrinck, B.A. and Shadduck, J.A., 1993. Ribosomal DNA sequences of Encephalitozoon hellem and Encephalitozoon cuniculi: species identification and phylogentic construction. J. Eukaryot. Microbiol. 40, 354-362.
Weiss, L.M. and Vossbrinck, C.R., 1999. Molecular biology, molecular phylogeny, and molecular diagnostic approaches to the microsporidia. In Wittner,M. and Wiss,L.M.(eds.), The Microsporidia and Microsporidiosis. ASM Press, Washington, D.C., pp. 129-171.
Williams, B.A.P., Hirt, R.P., Lucocq, J.M. and Embley, T.M., 2002. A mitochondrial remnant in the microsporidian Trachipleistophora homins. Nature 418, 865-869.
Wuyts, J., de Rijk,P., van de Peer,Y, Winkelmans,T., de Wachter,R., 2001. The European large subunit ribosomal RNA database. Nucleic Acids Res. 29, 175-177.
FIGURES
Fig. 1. Schematic diagram of Nosema bombycis rRNA gene. Mature rRNA gene domains are boxed. Locations of Primers on rRNA are given in Table 1. The arrows represent the 3’-end of each primer, the main parts of rRNA are produced by the gray-shaded primer sets. HG4R-c and HG4F-c are the
Fig. 2 The secondary structure model of Nosema bombycis for large subunit (LSU) rRNA. Helix numbering is according to de Rijk et al. (1998). The regions of the structure, where large variations between eukaryotic rRNAs are found, are boxed and labeled V1 to V12.
10 kb 8 kb 6 kb 5 kb 4 kb 3 kb 2.5 kb 2 kb 1.5 kb 1 kb 0.75 kb 8 kb 0.5 kb 10 kb 6 kb 5 kb 4 kb 3 kb 2.5 kb 2 kb 1.5 kb 1 kb 0.75 kb 0.5 kb
M
1
(A)
M 1 2 3 4
(B)
Fig. 3 The confirmation of Nosema bombycis rRNA organization. (A) Total length of rRNA was amplified with primer set, the amplicon (4,401 BP) containing all the rRNA sequence was yielded with a primer set, LSUF and 5SR. M: 1kb DNA ladder (Promega). (B) Amplification of internal fragments of rRNA. Lane 1: 3’end of LSUrRNA- ITS-main part of SSUrRNA (1,261 bp) by a primer set, ILSUF and HG4F-c. Lane 2: 3’end of LSUrRNA- ITS- SSUrRNA (1,700 bp) by a primer set, ILSUF and 1537R. Lane 3: 3’end of LSUrRNA- ITS- SSUrRNA-IGS-5S (2,093 bp) by a primer set, ILSUF and 5SR. Lane 4: main part of LSUrRNA-ITS-main part of SSUrRNA (3,031 bp) by a primer set, HG4R-c and HG4F-c. M: 1kb DNA ladder (Promega).
Table 1 Primers used for amplification and sequencing of Nosema bombycis rRNA.
Primer Sequence Amplicon Reference size (bp)
Large subunit rRNA (LSU) 2,108
LS228F 5’-GGA GGA AAA GAA ACT AAC-3’ Vossbrinck ILSUR 5’-ACC TGT CTC ACG ACG GTC TAA AC-3’ et al. 1993
5’ end of LSU
LSUF 5’-ACT CTC CTC TTT GCC TCA ATC A-3’
Internal transcribed spacer (ITS) 501
ILSUF 5’-TGG GTT TAG ACC GTC GTG AG-3’ S33R 5’-ATA GCG TCT ACG TCA GGC AG-3’
Small subunit rRNA (SSU) 1,232
18f 5’-CAC CAG GTT GAT TCT GCC-3’ 1537r 5’-TTA TGA TCC TGC TAA TGG TTC-3’
Intergenic spacer (IGS) and 5S rRNA 852
HG4F 5’-GCG GCT TAA TTT GAC TCA AC-3’ Gatehouse and
Malone, 1998
5SR 5’-TAC AGC ACC CAA CGT TCC CAA G-3’
Nosema bombycis putative pseudogene
KAI01N 5’-GTA GTA GAG ACC CAA ATA TC-3’ Kawakami etal., KAI02N 5’-ACT GTT CAG ATA TGG TCC TTA TCG-3’ 1994
(modified from KAI01 and KAI02, take the restrict enzyme site off)