National Chiao-Tung University PhD Dissertation 國立交通大學博士學位論文
Regulation of Two-component Systems in the Polymyxin B
Resistance and Capsular Polysaccharide Biosynthesis in Klebsiella
pneumoniae CG43
克雷白氏肺炎桿菌
CG43 雙分子系統與多黏菌素 B 抗性
及莢膜多醣體生合成之調控
生物科技學系 博士班 學生:鄭新耀(9229506) Student:Hsin-Yao Cheng 指導教授:彭慧玲博士 Advisor:Hwei-Ling Peng, Ph. D. 中華民國九十九年九月 September, 2010謝誌 (Acknowledgement)
博士班生涯結束的此刻,並沒有預期的喜悅,也許稍稍被迎接未來挑戰的壓力沖淡了,但我想更多 是內心深處的捨不得。這個結束象徵著另一個開始,在我跨步邁進之前,僅以生澀已久的字語,為這段 永難磨滅的記憶寫下註腳。 研究的過程往往幽暗未知,而無數良師益友們的鼓勵與支持,讓我有繼續走下去的勇氣。這些年來 始終最感激的,還是指導教授彭慧玲老師。學識上,彭老師提供豐富的資源與寬廣的空間,使我彷彿感 到自己沒有極限;彭老師更替我留心相關研討會訊息,使我有機會認識許多同領域甚至不同領域的學者, 拓展研究思考的觀點。生活上,彭老師親切地帶我度過研究生涯的適應期,躊躇時指引我研究的方向, 犯錯時不吝寬容的原諒,心情低落時帶我走出悲傷的氛圍。彭老師在經濟方面的協助,使我可以心無旁 騖的進行研究,學術論文與研究計畫書撰寫上,更是仔細引導,讓駑頓的我逐步摸索,修正錯誤。在學 術與生活上,我何其幸運可以接受彭老師的指導,老師謝謝您! 感謝清大分醫所的張晃猷老師,張老師淵博的學識與豐富研究經驗,一直在我的研究生涯中扮演不 可或缺的助力,論文及計畫書寫作的指導更讓我獲益良多,擔任資格考、非論文資格考及畢業口試委員 時,總能確切地指出我的不足之處,讓我在深入探討課題時能有適當的反思,張老師學者的風範將是我 研究生涯努力的目標。此外更要感謝張老師實驗室學姊與學弟妹持續的關心與支持。謝謝在中山醫學大 學任教的怡琪學姊,提供實驗設計與論文寫作上的最佳楷模,感謝親切與我討論實驗並熱心提供協助的 婕琳、莉芳與韻如學姊,此外實驗室學弟妹蕙如、Manish、幸瑜、漢聲與欣瑜,你們的陪伴使我獲得堅 持的勇氣。 感謝生科所楊昀良老師,大學時修習楊老師的課程,便感受到楊老師深厚的學識與獨特的親和力。 直至研究所資格考、非論文資格考及畢業口試,這份親和力伴隨深入中肯的見解,不僅消弭報告的緊張 感,更使我融入學術討論的樂趣。論文的仔細修改與列舉的小問題,使我體認楊老師細心嚴謹的治學態 度,並對往後的生涯有更高的期許。 感謝生科所林志生老師,林老師的課程自大學起就使我獲益良多,研究所時互動的上課模式與課堂 中的腦力激盪,使我無形中增長許多知識。研究所資格考、非論文資格考及畢業口試,林老師以各種觀 點給予的寶貴建議,不僅豐富論文,更拓展研究的格局。林老師鉅細靡遺地閱讀論文,並對於資料呈現 的格式提供許多建議,使我得以改善論文中諸多錯誤,對此深表感謝。除此之外,林老師對於學術環境 現實面的懇切提醒,也讓我對未來的挑戰有更深入的體認與心理準備。我瞭解並會永遠珍惜老師的諄諄 教誨,老師謝謝您! 感謝中興大學鄧文玲老師,除豐富的微生物學與分子技術的背景外,鄧老師在討論時讓我感覺更像 親切的實驗室學姊。在口試場合對於實驗結果的討論,往往讓我學得比報告前更多,特別是爽朗的笑聲 與機敏的反應,在談笑風生之間增長我的見聞。鄧老師熱情與謹慎並存的態度,與論文修改的貼心提醒, 讓我體會懂得並享受研究的樂趣。 感謝中研院陳金榜老師,陳老師在蛋白質結構的豐富研究成果,讓我獲益良多。同時也感謝陳老師 實驗室的博士班研究生羅世奇,在多次的討論中,使我對 PmrD 蛋白質結構與功能相關性有更進一步的認識,並激發許多研究的靈感。 在新竹的實驗室曾幾何時成了我第二個家,感謝這難得的緣分讓我認識實驗室的大家。感謝盈璁學 長,認真細心地指導還是專題生、懵懂的我,為我的研究生活鋪下第一塊磚;感謝定宇、平輝學長在剛 進入實驗室的初期,耐心教導我實驗技巧;感謝靖婷學姊帶領我進入雙分子系統的研究領域,也讓我熟 悉分子生物學的工具;感謝盈蓉學姊看著我完成大學部專題到博士班畢業,無論實驗技巧、思考邏輯、 寫作要領,都在學姊身上學到很多,學姊散發的活力與熱情,總是能讓四周的人快樂起來;婉君學姊爽 朗的笑聲與熱心的指導;珮瑄學姊美妙的歌喉、好吃的義大利麵料理與驚人的工作量;騰逸學長,怡欣、 巧韻、致翔學姊等親切的照顧,都是我充滿感恩的記憶;從大學開始就是同學的祐俊,那黝黑外表下的 細心讓我記憶深刻;認識十一年的健誠,除了驚人歌喉、細緻的陶藝巧手,更在許多實驗討論中與我分 享嶄新的想法,在實驗設計操作上的嚴謹態度與整理報告的功力令我望塵莫及;聰明努力的智凱,與我 分享許多小說心得、認真踏實的育聖、兼具美貌與智慧的心瑋;酷酷的南台灣帥哥格維、與我一起展開 暑假游泳減肥計畫的登魁、內向的朝陽;溫柔中帶著堅強的實驗室生力軍靜柔、外表粗獷做起實驗又格 外細心的秉熹、運動型的正妹研究生嘉怡、氣質彬彬頗有骨感的承哲;剛進實驗室時幾乎被我拎在身上 的顗峰與雅雯,看著你們成長進步,身為直屬學長的我倍感欣慰;努力學習的志桓,你認真的態度是每 個人的榜樣、優雅如貴婦的純珊,讓我知道新竹有很多好吃的簡餐店;甜美中帶點鄉土味的佩君、剛柔 並濟認真負責的家華,直升博班的哲充;網拍美少女品瑄與崴云、看起來總是有實驗後疲態的豪君;眼 神中帶點憂鬱的舉豪、譽為彭家趙又廷的帥哥力成、圓圓肚子漸漸消失的郁勝;專題生佳融、宗穎、冠 維、Stephanie (You’ll become really somebody!),你們都是我難以割捨的回憶。
謝謝吳東昆老師實驗室的程翔學長、媛婷學姊在實驗上的照顧與幫忙,以及最後論文撰寫的建議; 謝謝晉豪、晉源,實驗上慷慨的協助。謝謝廖光文老師實驗室的昱丞學弟,畢業的志豪、國領學長陪我 打球聊天,感謝同樣身為博士班研究生的存操、羿喬,彼此互相的鼓勵打氣。謝謝所辦小姐淑卿、佳文、 聖鈴、珍佑長久以來的幫忙。 謝謝我的十幾年來的好朋友仕賢、建宏、威任、佳穎、佩芳、佳翰、柄宏,在我失意的時候陪我喝 酒聊天,看電影吃美食抒發情緒,讓我可以適當轉換心情,繼續進行研究。 最後要感謝我的家人,在天堂的奶奶與外婆,我多麼希望與妳們分享這份喜悅;感謝我的老爸老媽, 還有可愛活潑的妹妹伊芳,如果沒有你們的全力支持,我想我無法走到這裡。你們的支持像一股不滅的 能量,使我努力的小小火花化為燦爛的光芒。僅以這微小的榮耀,獻給永遠難以替代的你們。 新耀 2010/9/28
論文摘要
克雷白氏肺炎桿菌是臨床上一種重要的病原菌,常造成伺機性感染,包含尿道感 染、肺炎、化膿性感染、原發性肝膿瘍及敗血症等。隨著多重抗藥菌株的產生與散佈, 改善目前治療方式,並深入瞭解其致病機轉顯得刻不容緩。為適應環境改變,病原菌常 藉由雙分子系統調控並適當表現其毒性因子。雙分子系統 (two-component system; 2CS) 由位於細胞內膜的感應蛋白及細胞質內的反應調控蛋白組成。本論文著重探討克雷白氏 菌中雙分子系統PhoP/PhoQ、PmrA/PmrB、RstA/RstB 及 RcsCDB 於其毒性因子的調控。 多黏菌素為近年重新評估使用的藥物之一,沙門氏菌中PhoP/PhoQ, PmrA/PmrB 及 PmrD 蛋白質共同調控多黏菌素 B 之抗性,為瞭解克雷白氏菌中多黏菌素 B 抗性的機 轉,我們對可能參與的基因進行突變。結果顯示,相對於參與莢膜多醣體生合成基因, 合成PmrF、PhoP、PmrA、PmrD 等參與脂多醣修飾及調控蛋白之基因缺損,會使克雷 白氏菌多黏菌素 B 抗性有較顯著的降低。藉由 LacZ 報導基因及 DNA 電泳遲滯實驗(electrophoretic mobility shift assay),我們發現 pmrD 基因表現受到 PhoP 的直接活化, 細菌雙雜合實驗 (bacterial two-hybrid analysis) 也證明 PmrD 與 PmrA 在細菌體內的交互 作 用 , 另 外 藉 由 磷 酸 根 傳 遞 分 析 (phosphotransfer assay) 與 激 酶 / 去 磷 酸 酶 實 驗 (kinase/phosphatase assay) 更指出,此交互作用可防止感應蛋白 PmrB 對 PmrA 的去磷酸
化。這些結果顯示克雷白氏菌 PmrD 扮演了連結雙分子系統 PhoP/PhoQ 及 PmrA/PmrB
的角色。進一步藉由點突變方式,我們在15 個所選擇的 PmrD 殘基中,發現 9 個改變
為丙氨酸後對多黏菌素抗性有顯著影響,顯示這些氨基酸序列可能在PmrD 與 PmrA 交
互作用扮演重要角色。
為瞭解克雷白氏菌RstA/RstB 之調控,我們利用 LacZ 報導基因證明 PhoP 及 RstA
可正向調控 rstA 基因表現。此外利用克雷白氏菌野生株與 rstA 基因缺損株 cDNA 進行 刪除雜交法 (subtractive cDNA hybridization),我們搜尋到 11 個可能受到 RstA 活化及 19 個可能受到 RstA 抑制的基因,結果顯示 RstA/RstB 可能參與調控鐵分子的運送、攝 取以及重金屬鉛之抗性;然而針對野生株、rstA、rstB 以及 rstArstB 基因缺損突變株進 行表型比較,顯示 rstA 或 rstB 基因缺損對於細菌在含鐵或缺鐵環境的生長、螯鐵分子 生合成 (siderophore biosynthesis)、重金屬鉛抗性、酸性環境適應性或膽鹽抗性均沒有顯 著影響,顯示克雷白氏菌中RstA/RstB 的功能仍待釐清。 除雙分子系統 RcsCDB 之外,克雷白氏菌 K2 莢膜多醣體生合成亦受到轉錄因子
RmpA2 的調控。在克雷白氏菌 CG43 大型毒性質體 pLVPK 序列中,我們發現另一個黏 性分子基因 rmpA。分析 rmpA 基因缺損株與 rmpA 回補菌株,我們發現 RmpA 可能與 RcsB 共同活化莢膜多醣體生合成。藉由細菌雙雜合實驗及免疫共沈澱分析,顯示 RmpA 與可與RcsB 交互作用。此外藉由 LacZ 報導基因、DNA 電泳遲滯實驗、有限稀釋反轉 錄聚合酶連鎖反應 (limiting-dilution RT-PCR) 及 fur 基因缺損株表型的分析,我們發現 rmpA 基因表現受到鐵離子攝取調控蛋白 Fur 抑制,顯示莢膜多醣體生合成與鐵離子攝 取在克雷白氏菌致病過程中可能存在交互作用。 綜合以上所述,本論文探討克雷白氏菌雙分子系統兩種調控方式:一種可藉由連結 蛋白PmrD 與反應調控蛋白 PmrA 相互作用,達成其轉譯後修飾來活化下游多黏菌素 B 抗性基因表現;另一種則包含反應調控蛋白RcsB 與其協助分子 RmpA 之共同作用,藉 以活化莢膜多醣體生合成。這些結果暗示在不同環境下,克雷白氏菌可藉由許多有彈性 及效率的方式調控其致病因子表現。
Thesis Abstract
Klebsiella pneumoniae, an important opportunistic pathogen, causes urinary tract infections, pneumonia, purulent infections, primary liver abscess and septicemia. The emergence and wide-spread of multiresistant strains has urged the improvement of current therapeutic strategies and a deeper understanding of its pathogenesis. Pathogenic bacteria usually utilize two-component systems (2CSs), consisting of an inner membrane sensor kinase and a cytoplasmic response regulator, to modulate the expression of virulence factors in response to environmental stimuli. The objectives of this dissertation were to investigate the functional role and regulation of 2CSs PhoP/PhoQ, PmrA/PmrB, RstA/RstB and RcsCDB in K. pneumoniae virulence determinants.
The 2CSs PhoP/PhoQ, PmrA/PmrB and a small protein PmrD regulated Salmonella enterica resistance to polymyxin B, one of the reevaluated antimicrobial agents. To investigate the regulation of polymyxin B resistance in K. pneumoniae, mutation effects of the genetic determinants likely involved in the resistance were investigated. Compared with the deletion of the genes involved in capsular polysaccharide (CPS) biosynthesis, the deletion of pmrF, phoP, pmrA or pmrD encoding the proteins participated in the lipopolysaccharide modifications or regulation resulted in a significantly reduced resistance. Through LacZ reporter assay and DNA electrophoretic mobility shift assay (EMSA), we have found that the expression of pmrD was directly activated by PhoP. Bacterial two-hybrid analysis has indicated an in vivo interaction between PmrD and PmrA. As illustrated by in vitro phosphotransfer assay and kinase/phosphatase assay, such an interaction could protect PmrA from the dephosphorylation by its cognate sensor protein PmrB. The results suggest a role of Klebsiella PmrD in connecting 2CSs PhoP/PhoQ and PmrA/PmrB. By point mutation strategy, 9 of the 15 selected residues on PmrD were shown to be critically involved in the polymyxin B resistance implying these residues are required for PmrD/PmrA interaction.
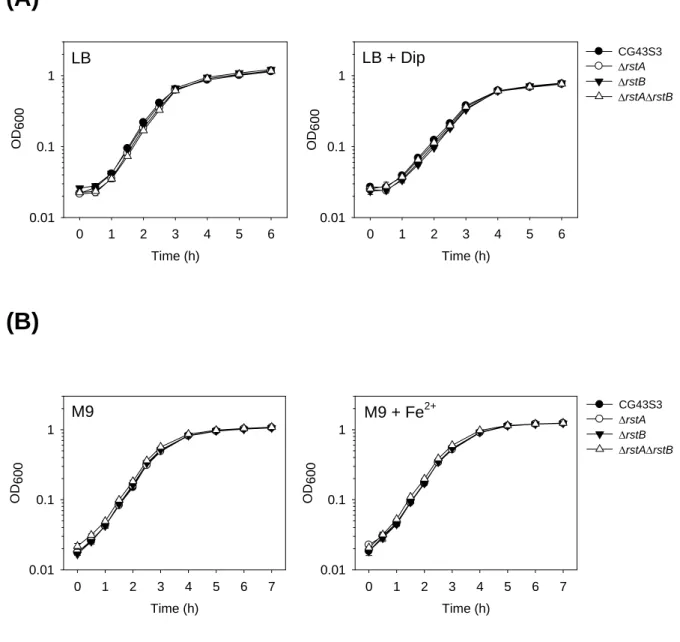
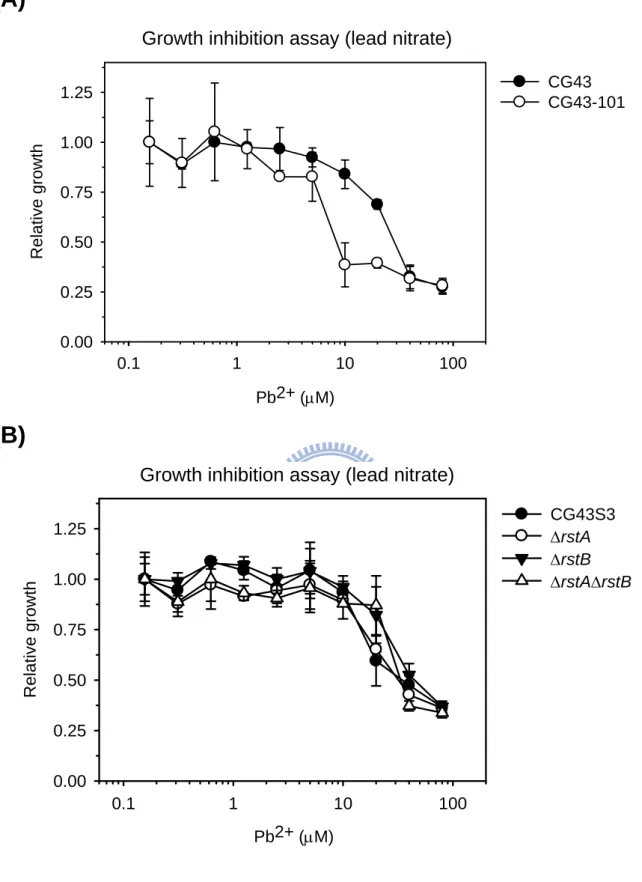
To characterize the 2CS RstA/RstB in K. pneumoniae, LacZ reporter assay conducted indicated a positive regulatory role of PhoP and RstA on the expression of rstA. Subtractive hybridization of the cDNA from the parental strain and isogenic ΔrstA mutant has led to the identification of 11 RstA-activated genes and 19 RstA-repressed genes involved in various cellular functions, such as iron transport/uptake and lead resistance. Comparative phenotype analysis of the wild-type strain, ΔrstA, ΔrstB and ΔrstAΔrstB mutants, however, showed that the deletion of rstA or rstB had no apparent effects on the growth under iron-depletion or
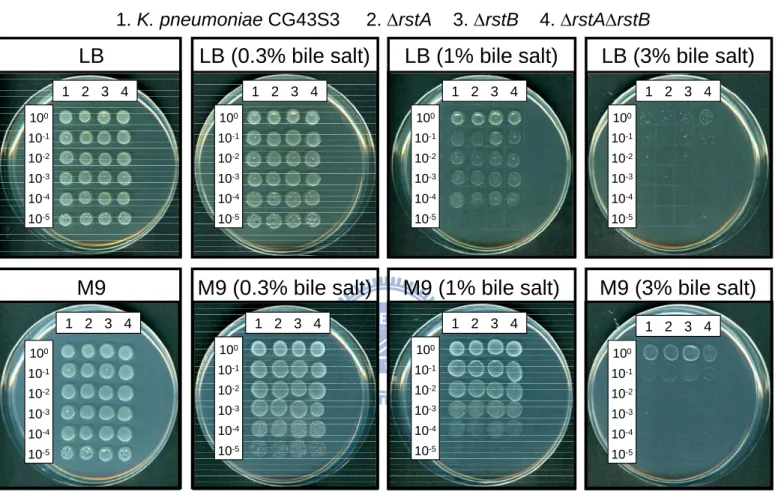
-repletion conditions, siderophore biosynthesis, lead resistance, acid stress response or the resistance to bile salts, indicating that the functional role of RstA/RstB in K. pneumoniae remained to be clarified.
In addition to the 2CS RcsCDB, K. pneumoniae K2 CPS biosynthesis was also regulated by the transcription factor RmpA2. Sequence analysis of pLVPK in K. pneumoniae CG43 has revealed rmpA, another mucoid factor encoding gene. Phenotype analysis of the rmpA deletion mutant and the complemented strain has implied that RmpA activated K2 CPS biosynthesis through an cooperation with RcsB. Both bacterial two-hybrid analysis and co-immunoprecipitation have further confirmed RmpA/RcsB interaction. Besides, through LacZ reporter assay, DNA EMSA, limiting-dilution PCR and phenotype analysis of Δfur mutant strain, we have shown that the expression of rmpA was negatively regulated by Fur, the global ferric ion uptake regulator. The results indicated the interplay between capsular polysaccharide biosynthesis and iron uptake in K. pneumoniae pathogenesis.
In summary, two modes of 2CS regulation in K. pneumoniae were investigated in this dissertation: one regarding the interaction between the connector protein PmrD and PmrA, resulting in a post-translational modification to enhance the expression of downstream genes involved in polymyxin B resistance; the other comprising the cooperation between the response regulator RcsB and its accessory factor RmpA for the activation of the K2 CPS biosynthesis. The findings indicated that under different environmental conditions, K. pneumoniae could regulate the expression of its virulence determinants in a versatile and efficient manner.
Table of Contents
謝誌 (Acknowledgement) ...i
論文摘要 ... iii
Thesis Abstract ...v
Table of Contents ...vii
List of Tables...xi
List of Figures...xii
Abbreviations ...xiv
CHAPTER 1 General Introduction ...1
1.1 Klebsiella pneumoniae...2
1.1.1 Virulence factors of K. pneumoniae ...2
1.1.2 Characteristics of K. pneumoniae infections ...2
1.2 Two-component systems (2CSs)...5
1.2.1 Classification of 2CSs...6
1.2.2 The sensor kinase...6
1.2.3 The response regulator...7
1.2.4 The 2CS network ...7
1.3 Thesis objectives...8
CHAPTER 2 Molecular Characterization of the PhoPQ-PmrD-PmrAB Mediated Pathway Regulating Polymyxin B Resistance in Klebsiella pneumoniae CG43 ... 11
2.1 Abstract...12
2.2 Introduction...13
2.3 Results ...15
2.3.1 Reduced production of capsular polysaccharide had minor effect on the polymyxin B resistance in K. pneumoniae ...15
2.3.3 Deletion effect of Klebsiella pmrA, pmrD or phoP on polymyxin B resistance ...16
2.3.4 Effect of pmrA, phoP or pmrD deletion on PpmrH::lacZ and PpmrD::lacZ expression ...17
2.3.5 Analysis of EMSA indicates a direct binding of the recombinant PhoP to PpmrD....17
2.3.6 Two-hybrid analysis of the in vivo interaction between Klebsiella PmrD and PmrA ...18
2.3.7 PmrD prevents the dephosphorylation of PmrA catalyzed by PmrB ...18
2.3.8 Identification of residues critical for PmrD functionality...19
2.4 Discussion...21
2.5 Figure ...23
CHAPTER 3 Functional Characterization of The Two-component System RstA/RstB in Klebsiella pneumoniae CG43...37
3.1 Abstract...38
3.2 Introduction...39
3.3 Results ...41
3.3.1 Regulation of rstA expression in K. pneumoniae CG43...41
3.3.2 Identification of RstA-regulated genes by subtractive cDNA hybridization...41
3.3.3 Sequence analysis of RstA-regulated genes ...42
3.3.4 Effect of iron on the growth of K. pneumoniae strains in rich and minimal medium ...43
3.3.5 Effect of lead on the growth of K. pneumoniae strains ...43
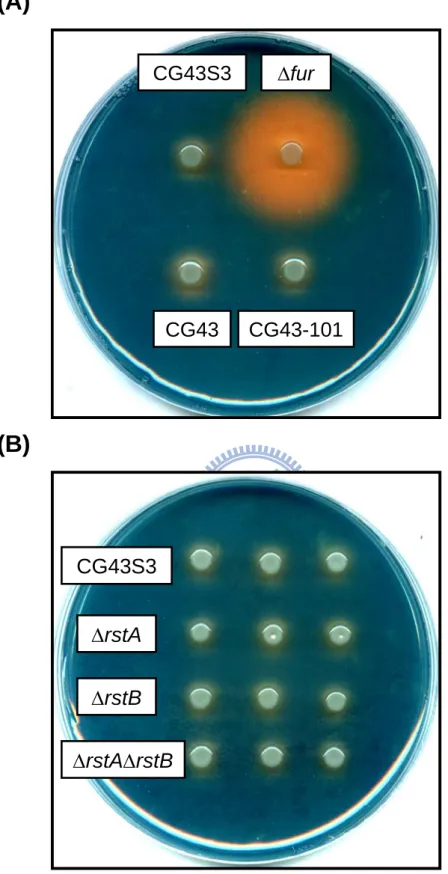
3.3.6 Siderophore biosynthesis of K. pneumoniae strains ...44
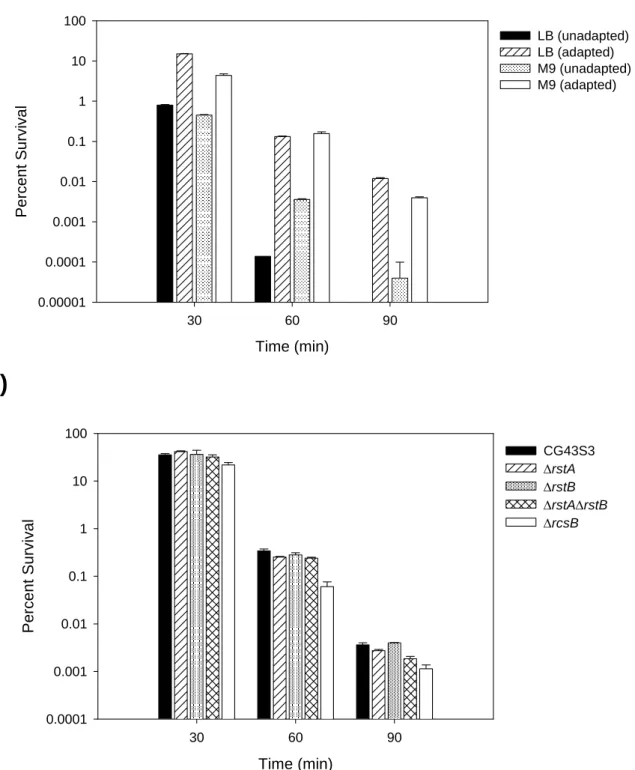
3.3.7 Acid tolerance response of K. pneumoniae strains ...44
3.3.8 Bile salt resistance of K. pneumoniae strains ...45
3.4 Discussion...46
3.5 Table ...48
3.6 Figure ...50
CHAPTER 4 RmpA Regulation of Capsular Polysaccharide Biosynthesis in Klebsiella pneumoniae CG43 ...58
4.2 Introduction...60
4.3 Results ...62
4.3.1 Comparison of rmpA/rmpA2 containing regions in K. pneumoniae CG43 ...62
4.3.2 Deletion of rmpA reduced CPS production and virulence...62
4.3.3 RmpA acted as an activator of cps expression ...63
4.3.4 Effect of poly(G) tract variation on rmpA/rmpA2 expression ...63
4.3.5 RmpA regulates cps expression in an RcsB-dependent manner...64
4.3.6 Interaction between RmpA and RcsB using two-hybrid analysis ...65
4.3.7 Co-immunoprecipitation analysis of the interaction between RmpA and RcsB ...66
4.3.8 The expression of rmpA is subjected to negative regulation by Fur...66
4.3.9 The recombinant Fur was able to bind specifically to PrmpA...67
4.3.10 Identification of rmpA transcriptional start site ...67
4.3.11 Deletion of fur led to overproduction of CPS...68
4.4 Discussion...69
4.5 Table ...73
4.6 Figure ...74
CHAPTER 5 Conclusion and Perspectives ...98
CHAPTER 6 Experimental Section ...102
6.1 Materials ...103
6.1.1 Plasmids, bacterial strains, primers and growth conditions...103
6.2 General Experimental Procedures ...104
6.2.1 Construction of specific gene-deletion mutants ...104
6.2.2 Extraction and quantification of CPS ...104
6.2.3 Polymyxin B resistance assay...105
6.2.4 Cell line, cell culture and phagocytosis assay ...105
6.2.5 Construction of reporter fusion plasmids and the measurement of promoter activity ...106
6.2.6 Cloning, expression and purification of recombinant proteins...106
6.2.7 DNA electrophoretic mobility shift assay (EMSA)...107
6.2.8 Bacterial two-hybrid assay ...108
6.2.10 Kinase/phosphatase and autokinase assay ...109
6.2.11 Construction of complementation plasmids encoding PmrD with point mutations ...109
6.2.12 Subtractive cDNA hybridization...110
6.2.13 Measurement of bacterial growth in iron depletion/repletion conditions...110
6.2.14 Growth inhibition assay... 111
6.2.15 Preparation of CAS agar plates... 111
6.2.16 Acid tolerance response ...112
6.2.17 Bile salt resistance ...112
6.2.18 Mouse lethality assay...113
6.2.19 Bacterial survival in serum ...113
6.2.20 Construction of the plasmid for K2 cps Porf1-2::lacZ chromosomal fusion...113
6.2.21 Preparation of RcsB-His antiserum ...113
6.2.22 Construction of GST fusion plasmids and co-immunoprecipitation ...114
6.2.23 Identification of rmpA transcriptional start site ...115
6.2.24 Limiting-dilution RT-PCR...115 6.2.25 Statistical analysis...116 6.3 Table ... 117 References ...130 Publication ...149 Vita ...150
List of Tables
Table 1.1. Risk factors associated with abscess formations in K. pneumoniae infections ...10
Table 3.1. RstA-activated genes identified by subtractive cDNA hybridization ...48
Table 3.2. RstA-repressed genes identified by subtractive cDNA hybridization ...49
Table 4.1. Virulence properties of K. pneumoniae strains. ...73
Table 6.1. Bacterial strains used in this study...117
Table 6.2. Plasmids used in this study ...119
List of Figures
Fig. 2.1. Deletion effects of ugd, wza and rcsB genes on K. pneumoniae K2 CPS production
and resistance to polymyxin B...24
Fig. 2.2. Involvement of K. pneumoniae pmrF gene in polymyxin B resistance and intra-macrophage survival ...25
Fig. 2.3. Effects of K. pneumoniae pmrA, pmrD and phoP deletion and complementation in polymyxin B resistance and intra-macrophage survival...27
Fig. 2.4. Schematic representation of pmrH and pmrD loci and determination of K. pneumoniae PpmrH::lacZ and PpmrD::lacZ activity ...29
Fig. 2.5. Binding of His-PhoP and His-PhoPN149 to PpmrD...31
Fig. 2.6. Klebsiella PmrD interacts with PmrA to prevent dephosphorylation...33
Fig. 2.7. Identification of critical residues involved in Klebsiella PmrD functioning...35
Fig. 2.8. A model illustrating the regulation of polymyxin B resistance in K. pneumoniae by PhoP/PhoQ, PmrD and PmrA/PmrB ...36
Fig. 3.1. Schematic representation of K. pneumoniae rstA locus and PrstA::lacZactivity measurements...51
Fig. 3.2. Identification of RstA-regulated genes by subtractive cDNA hybridization...52
Fig. 3.3. Growth of K. pneumoniae strains under iron-depletion/repletion...53
Fig. 3.4. Effect of lead on the relative growth of K. pneumoniae strains ...54
Fig. 3.5. Siderophore production of K. pneumoniae strains on CAS agar plates ...55
Fig. 3.6. Survival of K. pneumoniae strains after acid challenge ...56
Fig. 3.7. Growth of K. pneumoniae strains on bile salt-containing medium ...57
Fig. 4.1. Comparison of rmpA and rmpA2 containing PAI-like regions...74
Fig. 4.2. Comparison of precipitation speeds and K2 CPS production in K. pneumoniae strains ...76
Fig. 4.3. Expression of K2 cps genes in various genetic backgrounds...77
Fig. 4.4. Effect of poly(G) tract variation on RmpA and RmpA2 coding sequence...79
Fig. 4.5. RmpA and RcsB activated the expression of K2 cps genes in a coordinated manner ...80 Fig. 4.6. Bacterial two-hybrid analysis of the interaction between RcsA/RcsB, RcsB/RmpA,
and RcsB/RmpA2 proteins ...82 Fig. 4.7. Co-immunoprecipitation analysis of the interaction between RcsA/RcsB RcsB/RmpA,
and RcsB/RmpA2 proteins ...85 Fig. 4.8. Time course analysis of PrmpA::lacZ and PrmpA2::lacZ expression ...86
Fig. 4.9. Effect of fur deletion or iron depletion on the activity of PrmpA::lacZ, PrmpA2::lacZ,
PiucA::lacZ, and PiroB::lacZ ...89
Fig. 4.10. EMSA of the recombinant His-Fur and its target promoters...92 Fig. 4.11. Identification of rmpA transcription start site by 5’-RACE...94 Fig. 4.12. Phenotype comparison of K. pneumoniae CG43S3, fur deletion mutant, and the
complemented strain ...96 Fig. 4.13. A model illustrating the regulation of Klebsiella K2 CPS biosynthesis by RcsB and
its auxiliary factors RcsA, RmpA and RmpA2 and the regulation of rmpA expression by Fur ...97
Abbreviations
2CS(s) two-component system(s)
bp(s) base pair(s)
AMV avian myeloblastosis virus AP(s) antimicrobial peptide(s) CPS capsular polysaccharide CFU colony forming unit
EDTA ethylenediaminetetraacetic acid ESBL extended-spectrum β-lactamase
EMSA electrophoretic mobility shift assay HDTMA hexadecyltrimethylammonium bromide Hpt histidine-containing phosphotransfer
HV hypermucoviscosity
IPTG isopropyl 1-thio-β-D-galactopyranoside
kb kilobase(s) kDa kilodalton(s) LB Luria-Bertani μg microgram mL mililiter μL microliter mM milimolar μM micromolar
MOPS 3-(N-morpholino)hpropanesulfonic acid
ng nanogram
OD optical density
ONPG ο-nitrophenyl-β-D-galactopyranoside
PAGE polyacrylamide gel electrophoresis PAI pathogenecity island
PIPES piperazine-N,N’-bis(2-ethanesulfonic acid) PLA pyogenic liver abscess
rpm revolutions per minutes RT reverse transcriptase SDS sodium dodecyl sulfate
CHAPTER 1
1.1 Klebsiella pneumoniae
Klebsiella pneumoniae is a rod-shaped, non-motile, heavily-encapsulated gram-negative bacterium belonging to the Enterobacteriaceae. The Klebsiella spp. are ubiquitous in nature, with two plausible habitats: one is the surface water, soil, sewage and the plants (9, 25, 58, 157, 212), and other is the mucosal surfaces of animal hosts including humans, whereas two common habitats in humans are nasopharynx and the intestinal tract (196).
1.1.1 Virulence factors of K. pneumoniae
The factors contributing to K. pneumoniae pathogenesis include capsular polysaccharide (CPS), lipopolysaccharide (LPS), iron acquisition systems and adhesion factors. Clinically isolated K. pneumoniae usually produces large amounts of capsular polysaccharide (CPS) and therefore forms large glistering colonies with viscid consistency. As a major virulence factor, CPS acts to protect the bacteria from phagocytosis, from killing by polymorphonuclear granulocytes and from killing by bactericidal serum factors (62, 70, 143). Besides the physical hindrance to fimbrial binding, the role of Klebsiella CPS in mediating the bacterial resistance to antimicrobial peptides has also been reported (30, 150). K. pneumoniae strains expressing K1 and K2 CPS are the most virulent to mice (167). In addition to the 77 serotypes distinguished, a new K serotype has been identified recently (186). Most recently, the hypermucoviscosity (HV) phenotype of K. pneumoniae isolates resulting from a profound expression of CPS has also been correlated with the development of invasive syndrome (137).
The LPS O-antigen and the lipid A are responsible for the resistance to serum factors and the establishment of septic shock (62, 160). The adhesion factors, including the type 1 (223, 225, 226) and 3 pilus (107, 131, 222) and the non-fimbrial adhesion CF29K (50) and KPF28 (51, 52), were associated with the initial attachment and subsequent colonization of K. pneumoniae in the respiratory and urinary tract. The aerobactin-mediated iron uptake system, which has been reported to be located in a large resident plasmid, also contributed to the virulence of K. pneumoniae (48, 177, 237).
1.1.2 Characteristics of K. pneumoniae infections
As an opportunistic pathogen, the vast majority of K. pneumoniae infections were found in immuno-compromised individuals who are hospitalized and suffered from severe underlying diseases, such as diabetes mellitus or chronic pulmonary obstruction. In respect of
bacteremia caused by nosocomial gram-negative pathogens, Klebsiella is second only to Escherichia coli (270). Reported carrier rates in hospitalized patients are 77% in the stool, 19% in the pharynx and 42% on the hands of patients (47, 213). Such high rates of nosocomial colonization, however, were likely due to the use of antibiotics rather than the factors with delivery of care in the hospital (198, 207). The widespread use of antimicrobial therapy has been implicated to be responsible for the occurrence of multiresistant strains of K. pneumoniae in hospitals (213, 242) during outbreaks of nosocomial infections since 1980s (6, 75, 81, 252). The most common site of infection caused by K. pneumoniae is the urinary tract, which accounts for 6 to 17% of all nosocomial infections and shows an even higher incidence in patients with neuropathic bladders or with diabetes mellitus (15, 153).
Compared with other primary pathogens such as E. coli and Salmonella spp., K. pneumoniae has drawn less attention due to its opportunistic nature. However, with the extensive use of antibiotics and the widespread of multiresistant strain, especially the extended-spectrum β-lactamase (ESBL)-producing strains emerged since 1982 (71, 75, 159, 161), there has been renewed interest in K. pneumoniae infections. The resistance of K. pneumoniae to third-generation cephalosporins was usually conferred via the biosynthesis of ESBLs to hydrolyze broad-spectrum cephalosporins, monobactans and penicillins. In addition to increased frequency of β-lactam resistance, variations in the class of β-lactamase have emerged recently. The previously prevailing SHV and TEM enzymes were replaced by CTX, and strains producing CTX-ESBLs often showed resistance to fluoroquinolones and ciprofloxacin (22). The notion that ESBLs are often plasmid-mediated and that multiresistant K. pneumoniae strains usually harbor a relatively stable plasmid encoding ESBL have rendered K. pneumoniae a major threat to the ever-increasing number of susceptible individuals. Initially started from Argentina, the strains have now also been observed in areas other than Latin America (172, 197, 201), including Europe and Asia (148, 181). ESBL-producing K. pneumoniae strains have thus become more important due to the associations with treatment failure, prolonged stay in the hospital, increased health expense and a possible increase in mortality (189). With the unusual pattern of antibiotic resistance of the organism, physicians were forced to use unconventional agents such as tigecycline, polymyxin, colistin and inhalatory aminoglycosides with limited success (124).
The community-acquired K. pneumoniae infections were often treated empirically according to the prevalence of antimicrobial resistance. Risk factors for ESBL-producing K. pneumoniae included previous hospitalization, antimicrobial used in the 3 months, age over
60, male gender and diabetes mellitus (124). Although varied from country to country, the emergence of infections from ESBL-producing strains has been reported in Europe, Asia, and South and North America (17, 44, 195). Though most strains produced TEM type ESBL, an increase in occurrence of CTX has been reported (152). Carbapenems are considered to be the preferred agents for the treatment of serious infections caused by ESBL-producing K. pneumoniae strains due to their stability to β-lactamase hydrolysis. However, strains capable of express carbapenemases of the KPC variety in nosocomial infections have been reported recently in America (178), Brazil (168), the United Kingdom (259) and Greek (240). Many are only susceptible to tigecycline and colistin while some have developed resistant as well (138), imposing a major challenge for public health.
During the last decade, K. pneumoniae infections causing community-acquired primary pyogenic liver abscess (PLA) have also become an emerging disease receiving increasing attention. Distinct from the classical PLA, which is a complication of intra-abdominal or biliary tract infections resulting from multiple aerobic and anaerobic bacterial strains (158), PLA caused by primary infection of K. pneumoniae as a single pathogen is often cryptogenic and often complicated with metastatic lesions (135, 147, 248). The strains belonging to K1 serotype was predominant, and most cases have been reported from Taiwan (42), which were associated with a distinct invasive syndrome in liver abscess, meningitis and endophthalmitis. Other cases have also been reported from other parts of Asia including Korea (43) and Singapore (236, 268) while there are also reports in other parts of the world including North America (68, 133, 199), Australia (243) and Europe (34, 115). The geographical diversity may have been due to interaction between bacterial and host variables, socio-economic factors and genetic susceptibility varied across racial groups (124).
Attempts have been made to identify the risk factors for K. pneumoniae infections associated with abscess-formation. As listed in Table 1.1, diabetes mellitus is the most tightly associated with K. pneumoniae PLA among the host factors. With respect to K. pneumoniae associated risk factors, phenotypic attributes like hypermucoviscosity, K1/K2 capsule serotype, and genetic factors such as magA and rmpA are the most frequently reported. To date, the correlation between HV phenotype and K. pneumoniae PLA, community-acquired urinary tract infections (146) and bacteremia (137, 271) has been generally accepted while he relationship between a specific K serotype, especially K1, and K. pneumoniae PLA was still debatable. The gene rmpA, which was originally isolated from a large resident plasmid in K. pneumoniae strains of K2 serotype (175), has been reported to encode the ability to enhance
the biosynthesis of exopolysaccharides in E. coli (176). RmpA2, encoded by a gene homologous to rmpA, has been shown to activate Klebsiella K2 CPS biosynthesis at the transcriptional level (130). The magA gene was originally identified from a tissue-invasive K1 strain NTUH-K2044 and was found to be present in all isolates of K1 serotype (65). Toll-like receptor 4 recognition has been implicated in the infection by K. pneumoniae strains carrying magA (263). Most recent finding indicated that magA should be renamed as wzyK1,
encoding the polymerase for K1 CPS biosynthesis (66, 269). In addition, the loci derived from pLVPK, a 219,385-bp plasmid of an invasive K2 strain CG43 (241), has also been implicated in the abscess formation during K. pneumoniae infections (237). A search in the genetic requirements of PLA in a mice oral infection model by K. pneumoniae has identified a set of regulatory genes suggesting the prerequisite formation of a regulatory network during the infection process (241). Despite these findings, the existence of other bacterial factors mediating K. pneumoniae pathogenesis, especially in PLA, remained elusive. Also, the widespread of multiresistant K. pneumoniae strains has urged the demands for the prevention of infections, rapid detection of infections and efficient treatments more urgent than before. As a result, a deeper understanding of the regulatory mechanisms of K. pneumoniae virulence factors would be required for future advances in the new therapeutic strategies.
1.2 Two-component systems (2CSs)
Bacteria must be able to sense and respond to their environment in order to survive. One major way microbes accomplish this is using signal transduction machinery such as two-component systems (2CSs). 2CSs are distributed widely throughout the prokaryotes and eukaryotes, with the exception of animals, rendering them as a target for the development of novel antimicrobial agents (85, 86).
As the most prevailing signal transduction mechanisms to control gene expression in bacteria, a typical 2CS consists of a membrane-associated sensor kinase and a cytoplasmic response regulator. In response to an external stimulus, the sensor protein undergoes auto-phosphorylation at a conserved histidine residue, and the signal is subsequently relayed to the cytosol via phosphorylation at the aspartic acid in the receiver domain of the response regulator (27). The signal transduction cascade may thus activate genes required for bacterial virulence during infections or survival in hostile environments (12). As is widely believed, 2CS proteins function as components of a signal transduction network, enabling bacteria to respond to complex environmental stimuli (5).
1.2.1 Classification of 2CSs
The 2CSs were classified into three major types by virtue of their functional domains: the classical system, the unorthodox system, and the hybrid system (204). Mostly, a 2CS sensor kinase carrying one input domain and one transmitter domain was called a classical (IT-type) sensors kinase. The others contain both the sensor kinase signature and a receiver domain of the response regulator, which were referred to as hybrid (ITR-type) sensors kinases. A small fraction of the hybrid sensors possesses an additional output domain at the carboxyl terminus and were referred to as unorthodox (ITRO-type) sensor kinases. For example, in P. aeruginosa PAO1, there are 42 IT-type sensor kinases, 12 ITR-type senor kinases, and 5 ITRO-type sensor kinases. Only three Hpt (histidine-containing phosphotransfer) modules had been observed (204). With the increasing number of bacterial 2CS identified, an emerging theme is the discovery of auxiliary factors, which are capable of influencing phosphotransfer but distinct from sensor kinases and response regulators. As the number of auxiliary factors increases, the members of a 2CS would surely increase, forming a three (and more) component system (27). Auxiliary factors could either exert its function by targeting the response regulator, as in the case of the unorthodox 2CS response regulators RcsB and its auxiliary factor RcsA (122, 155, 251). Auxiliary factors could also target on the sensor protein. One of the examples in a multiple component system is the Rap and Spo0E family phosphatases that interact with, and stimulate the auto-phosphatase activity of, the Spo0F and Spo0A response regulators respectively (219).
1.2.2 The sensor kinase
Each sensor kinase contains a variable input domain that is adapted for the detection of a specific stimulus, typically a chemical ligand, and a conserved transmitter domain that transfers the signal to its cognate response regulator through a phosphorylation cascade (57). A prototype sensor kinase is a homodimeric integral membrane domain in which the sensor domain is depicted as an extracellular loop within two membrane-spanning segments. The transmitter domain is localized within the cytoplasm. Recently the domains adapted by sensor proteins have been classified as extracellular sensor domains such as PDC (PhoQ-DcuS-CitA) (39, 40, 215), or membrane-embedded sensor domain similar to the phototaxis sensory rhodopsin II-transducer complex HtrII-SrII (84) and cytoplasmic sensor domains such as PAS (7, 123), GAF (102) or PHY (266). Although there is an apparent lack of uniformity between the specific conformational changes that take place within sensor domains upon changes in stimulus, the recent structural evidence suggests that the communication between
sensor and transmitter domains in sensor protein signaling is mediated by subtle structural changes along the dimmer interface and that related aspects of symmetry and asymmetry may also exist.
Signal termination in 2CSs usually occurs via the loss of the phosphoryl group from the response regulator. For example, the Spo0E family phosphatases (56, 88), which catalyze the dephosphorylation of the response regulator Spo0A involved in the initiation of endospore formation not only in Bacillus subtilis but also in other gram-positive bacteria, contain sequence and structural features similar to the chemotaxis phosphatases of CheZ (126, 275) and CheC/CheX/FliY families (188, 191, 230).
1.2.3 The response regulator
Consisted of a receiver domain and an output domain, the response regulator typically acts as the effector of 2CS. The input or receiver domain, which is usually conserved among regulators, received signals transferred from the sensor kinase and in some case from small phosphor donors such as phosphoramidates and acyl phosphates (258). Phosphorylation of the receiver domain is usually followed by a conformational change of the response regulator, which subsequently results in dimerization of the protein.
In contrast to the input domain, the output domain was rather diverse. Though most response regulators act as transcriptional regulators with various DNA-binding domains, the output domains of response regulators covered effector domains with a wide range of cellular functions, including RNA-binding, chemotaxis, two-component phosphorelay, cyclic di-GMP signaling, cAMP signaling and protein Ser/Thr phosphorylation (76). The properties that no restriction was applied on the variety of domains fused with the receiver domain and many 2CSs were able to interfere with the functioning of other signal transduction systems put 2CSs at the top of the bacterial signaling hierarchy.
1.2.4 The 2CS network
The increasing knowledge of 2CSs has revealed a diversity of designs controlling the flow of information within and between circuits. As a result, a 2CS network could be expected from two aspects, either the control of phosphorylation/dephosphorylation, or the signal integration and distribution among phosphotransfer pathways.
The control of phosphorylation/dephosphorylation covers single-step phosphotransfer, in which the sensor kinase exerts only its kinase activity, like the histidine kinase CheA in chemotaxis circuits (126), or exerts both kinase and phosphatase activity, as exemplified by
the E. coli 2CSs OmpR/EnvZ (11), PhoP/PhoQ (166) and CpxR/CpxA (217). In the latter case, a high expression level of the histidine kinase will increase the complex formation of the kinase/response regulator pair, with a commitment decrease in the response regulator phosphorylation.
The occurrence of phosphorelay in a 2CS could be found either in separate components, as in the sporulation phosphorelay of B. subtilis (180), or in hybrid proteins which usually contain reversible phosphorylation (4, 80) as also observed in RcsCDB phosphorelay regulating E. coli group I CPS biosynthesis (155). Possibly, the evolution of such complex phosphorelay machinery in 2CS signaling could provide additional points of control.
Finally, the autoregulation, which is resulted from a positive feedback from the 2CS itself, probably modulates the sensitivity to the applied stimulus, as explored in the BvgA/BvgS in Bordetella bronchiseptica (255). This may also create a short-term “learning” behavior, as shown in 2CS PhoB/PhoR in E. coli (103). A negative autoregulation could even give rise to an oscillatory behavior in the expression of downstream genes, as observed in the 2CS CovS/CovR in Streptococcus pyogenes (93).
In view of signal integration and distribution among phosphotransfer pathways, the 2CS network contains branched pathways of either “one-to-many” in the chemotaxis regulatory system (126), “many-to one” in the quorum sensing network of Vibrio harveyi (151), or “cross-regulation” via auxiliary proteins in S. enterica PmrD connector-mediated circuit (117, 119). Though much has been investigated in the complex 2CS network, a need remains for better understanding towards the properties of different circuit architectures to identify potential drug targets.
1.3 Thesis objectives
The prevalent presence of 2CS encoding genes in the bacterial genome as well as the wide spectrum of physiological functions governed by different 2CSs, such as virulence properties, have aroused our interest in studying the 2CSs in K. pneumoniae. Based on the available E. coli 2CSs listed in the KEGG database (http://www.genome.ad.jp/kegg/pathway/), a previous search of the homologous 2CS-coding genes in the genome sequence of K. pneumoniae MGH78578 (http://genome.wustl.edu/) has been performed. Among the 29 sensor kinase coding genes and 26 response regulator coding genes identified, the 2CSs PhoP/PhoQ, PmrA/PmrB, RstA/RstB and RcsCDB coding genes were also included. The role of 2CSs PhoP/PhoQ, PmrA/PmrB in S. enterica pathogenesis has been well documented (2, 13, 78, 205,
260). A novel mechanism connecting 2CSs PhoP/PhoQ and PmrA/PmrB by a small protein PmrD has shed light on the complex and versatile regulation of 2CS target genes (59, 117). RstA/RstB has been identified as a member of magnesium stimulon governed by the master 2CS PhoP/PhoQ (164). Recent studies have shed light its functional role in bacterial acid resistance (214), iron transport (111) and the survival against stressed encountered in the digestive tract (69). Despite these findings, the function of RstA/RstB remained rather limited. In addition, the RcsCDB phosphorelay has been reported to regulate E. coli group I CPS (87) and K. pneumoniae K2 CPS biosynthesis (245). In K. pneumoniae, in addition to RmpA2 which activated K2 cps expression independently to RcsB (130), another mucoid factor encoding gene rmpA was also indentified in pLVPK (37). Based on the critical role of CPS in K. pneumoniae pathogenesis, it would be of importance to investigate the mode of regulation of RmpA as well as its interplay with the RcsCDB phosphorelay. The objectives of this thesis are to investigate the functional role of PhoP/PhoQ, PmrA/PmrB, RstA/RstB and RcsCDB in the regulation of K. pneumoniae virulence determinants, and an overview is listed as follows:
Chapter 2 described the functional role of bacterial CPS as well as 2CSs PhoP/PhoQ and PmrA/PmrB in the regulation of polymyxin B resistance in K. pneumoniae CG43. In addition to the characterization of the PmrD connector-mediated pathway in K. pneumoniae at both genetic and the molecular level, the residues implicated in PmrD functioning were also explored via a mutagenesis approach.
Chapter 3 presents the characterization of the 2CS RstA/RstB in K. pneumoniae CG43. The involvement of PhoP and RstA on the expression of rstA was investigated, and RstA-regulated genes were identified by subtractive cDNA hybridization. Based on the results from cDNA subtraction and recent findings in RstA/RstB, phenotype analysis was conducted to explore the possible biological functions of RstA/RstB in K. pneumoniae.
Chapter 4 reports RmpA regulation of K. pneumoniae K2 CPS biosynthesis. The cooperation of RmpA and the RcsCDB phosphorelay was implicated from phenotype analysis and was demonstrated through both in vivo and in vitro approaches. A comparative analysis of RmpA and RmpA2 has further led to the identification of Fur, a global iron uptake regulator, in Klebsiella K2 CPS biosynthesis as well as the interplay of iron uptake and capsule biosynthesis in K. pneumoniae pathogenesis.
Chapter 5 concludes the entire thesis from a comprehensive point of view and provides perspectives regarding further investigations on the regulatory mechanisms and biological functions of the 2CSs herein investigated.
Table 1.1. Risk factors associated with abscess formations in K. pneumoniae infections
Risk factor Description Reference
K. pneumoniae associated factor
Phenotype attributes Hypermucoviscosity (HV) (136, 237, 272)
Extended spectrum β-lactamase (ESBL) productiona (136)
Capsular serotype (K1 or K2) (43, 67, 136, 143,
243, 267, 268) Genetic factors
K1-specific gene magA, allS (42, 68, 136, 154)
HV regulatory gene rmpA, rmpA2 (136, 272)
Integrative and conjugative element ICEKp1 (Homologue of Yersinia high-pathogenecity island (HPI)) (145)
Iron acquisition system iucABCEiutA, iroAiroNDCB, kfu (68, 272)
pLVPK derived loci terW+iutA+rmpA+silS+ (237)
Host factor
Underlying disease Diabetes mellitus (134, 237, 239)
Urinary tract obstruction (239)
Malignancy (239)
Association with focal infections Pneumonia (239)
Urinary tract infections (239)
Therapeutic treatments Poor glycemic control (144)
Disease acquisition Community acquisition (237)
a. Negatively correlated.
CHAPTER 2
Molecular Characterization of the PhoPQ-PmrD-PmrAB
Mediated Pathway Regulating Polymyxin B Resistance in
2.1 Abstract
The cationic peptide antibiotic polymyxin has recently been reevaluated in the treatment of severe infections caused by gram negative bacteria. In this study, the genetic determinants for capsular polysaccharide level and lipopolysaccharide modification involved in polymyxin B resistance of the opportunistic pathogen Klebsiella pneumoniae were characterized. The expressional control of the genes responsible for the resistance was assessed by a LacZ reporter system. The PmrD connector-mediated regulation for the expression of pmr genes involved in polymyxin B resistance was also demonstrated by DNA EMSA, two-hybrid analysis and in vitro phosphor-transfer assay. Deletion of the rcsB, which encoded an activator for the production of capsular polysaccharide, had a minor effect on K. pneumoniae resistance to polymyxin B. On the other hand, deletion of ugd or pmrF gene resulted in a drastic reduction of the resistance. The polymyxin B resistance was shown to be regulated by the two-component response regulators PhoP and PmrA at low magnesium and high iron, respectively. Similar to the control identified in Salmonella, expression of pmrD in K. pneumoniae was dependent on PhoP, the activated PmrD would then bind to PmrA to prolong the phosphorylation state of the PmrA, and eventually turn on the expression of pmr for the resistance to polymyxin B. The study reports a role of the capsular polysaccharide level and the pmr genes for K. pneumoniae resistance to polymyxin B. The PmrD connector-mediated pathway in governing the regulation of pmr expression was demonstrated. In comparison to the pmr regulation in Salmonella, PhoP in K. pneumoniae plays a major regulatory role in polymyxin B resistance.a
a A part of this chapter has been published:
1. Cheng, H. Y., Y. F. Chen, and H. L. Peng. 2010. Molecular characterization of the PhoPQ-PmrD-PmrAB mediated pathway regulating polymyxin B resistance in Klebsiella pneumoniae CG43. J Biomed Sci 17:60-75.
2. Luo, S. C., Y. C. Lou, H. Y. Cheng, Y. R. Pan, H. L. Peng, and C. Chen. 2010. Solution structure and phospho-PmrA recognition mode of PmrD from Klebsiella pneumoniae. J Struct Biol. (In press)
2.2 Introduction
Klebsiella pneumoniae, an important nosocomial pathogen, causes a wide range of infections including pneumonia, bacteremia, urinary tract infection, and sometimes even life-threatening septic shock (196). The emergence of multiresistant K. pneumoniae has reduced the efficacy of antibiotic treatments and prompted the reevaluation of previously but not currently applied antibiotics (63, 64) or a combined therapy (116). Polymyxins, originally isolated from Bacillus polymyxa, have emerged as promising candidates for the treatment of infections (274). As a member of antimicrobial peptides (APs), the bactericidal agent exerts its effects by interacting with the lipopolysaccharide (LPS) of gram-negative bacteria. The polycationic peptide ring on polymyxin competes for and substitutes the calcium and magnesium bridges that stabilize LPS, thus disrupting the integrity of the outer membrane leading to cell death (95, 274).
The Klebsiella capsular polysaccharide (CPS), which enables the organism to escape from complement-mediated serum killing and phagocytosis (53, 113), has been shown to physically hinder the binding of C3 complement (45) or polymyxin B (30). The assembly and transport of Klebsiella CPS followed the E. coli Wzy-dependent pathway (253), in which mutations at wza encoding the translocon protein forming the complex responsible for CPS polymer translocation and export resulted in an inability to assemble a capsular layer on the cell surface (55). The CPS biosynthesis in K. pneumoniae was transcriptionally regulated by the two-component system (2CS) RcsCDB (155) where the deletion of the response regulator encoding gene rcsB in K. pneumoniae caused a loss of mucoid phenotype and reduction in CPS production (130).
In Escherichia coli and Salmonella enterica serovar Typhimurium, polymyxin B resistance is achieved mainly through the expression of LPS modification enzymes, including PmrC, an aminotransferase for the decoration of the LPS with phosphoethanolamine (125) and the pmrHFIJKLM operon (92, 260) (also called pbgP or arn operon (20, 265)) encoding enzymes. Mutations at pmrF, which encoded a transferase for the addition of 4-aminoarabinose on bactoprenol phosphate, rendered S. enterica and Yersinia pseudotuberculosis more susceptible to polymyxin B (92, 156). The S. enterica ugd gene encodes an enzyme responsible for the supply of the amino sugar precursor L-aminoarabinose
for LPS modifications and hence the Ugd activity is essential for the resistance to polymyxin B (235). On the other hand, the E. coli ugd mutant with an impaired capsule also became
highly susceptible to polymyxin B (128).
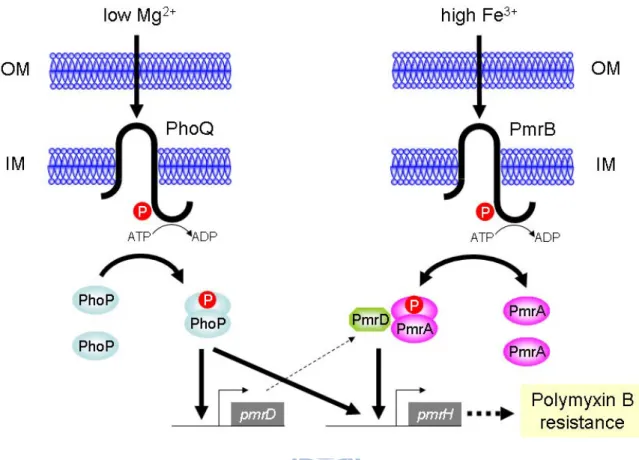
The 2CS PmrA/PmrB, consisting of the response regulator PmrA and its cognate sensor kinase PmrB, has been identified as a major regulatory system in polymyxin B resistance (91, 261). The resistance in S. enterica or E. coli has been shown to be inducible by the extracellular iron (256). In addition to acidic pH (193), the role of ferric ions as a triggering signal for the expression of PmrA/PmrB has been demonstrated (261). The 2CS PhoP/PhoQ which regulates the magnesium regulon (120) could also activate polymyxin B resistance under low magnesium in S. enterica, in which the PhoP/PhoQ-dependent control is connected by the small basic protein PmrD. The expression of pmrD could be activated by PhoP while repressed by PmrA forming a feedback loop (118, 127). The activated PmrD could then bind to the phosphorylated PmrA leading to a persistent expression of the PmrA-activated genes (117).
The PmrD encoding gene was also identified in E. coli and K. pneumoniae. However, pmrD deletion in E. coli had no effect on the bacterial susceptibility to polymyxin B (256). Recently, the PhoP-dependent expression of pmrD has also been demonstrated in K. pneumoniae. According to the predicted semi-conserved PhoP box in the pmrD upstream region, a direct binding of PhoP to the pmrD promoter for the regulation was speculated (165).
In this study, specific deletions of genetic loci involved in CPS biosynthesis and LPS modifications were introduced into K. pneumoniae CG43, a highly virulent clinical isolate of K2 serotype (36). Involvement of the genetic determinants in polymyxin B resistance was investigated.
2.3 Results
2.3.1 Reduced production of capsular polysaccharide had minor effect on the polymyxin B resistance in K. pneumoniae
K. pneumoniae CG43 is a highly encapsulated virulent strain (36). In order to verify the role of CPS in polymyxin B resistance, the Δugd and Δwza mutants were generated by allelic exchange strategy, and their phenotype as well as the amount of CPS produced were compared with the parental strain CG43S3 and ΔrcsB mutant (130). As shown in Fig. 2.1A, the Δugd and Δwza mutants formed apparently smaller colonies on LB agar plate compared with the glistering colony of the parental strain CG43S3. Although the colony morphology of the ΔrcsB mutant was indistinguishable from CG43S3, the CPS-deficient phenotype was evident as assessed using sedimentation assay and the amount of K2 CPS produced (Fig. 2.1B). Deletion of rcsB resulted in an approximately 50% reduction of the CPS, while the Δwza mutant produced less than 20% of that of its parental strain CG43S3. The CPS biosynthesis in Δugd mutant was almost abolished, indicating an indispensible role of Ugd in CPS biosynthesis. To investigate how the CPS level was associated with polymyxin B resistance, the survival rates of the strains challenged with polymyxin B were compared. The Δugd mutant producing the lowest amount of CPS was extremely sensitive to the treatment of polymyxin B (Fig. 2.1C). Although the Δugd mutant was CPS-deficient, the impaired polymyxin resistance may have been largely attributed to the defect in LPS biosynthesis since the survival rates of Δwza and ΔrcsB mutants appeared to be comparable with the parental strain CG43S3. This argues against the notion that the level of polymyxin B resistance is positively correlated to the amount of CPS (30). Nevertheless, the possibility that a higher amount of CPS was required for the resistance could not be ruled out. As shown in Fig. 2.1D, the introduction of pRK415-RcsB (38) resulted in a significantly higher resistance to polymyxin B in both ΔrcsB mutant and its parental strain. This indicated a protective effect of large amounts of CPS in polymyxin resistance.
2.3.2 PmrF is involved in polymyxin B resistance and survival within macrophage
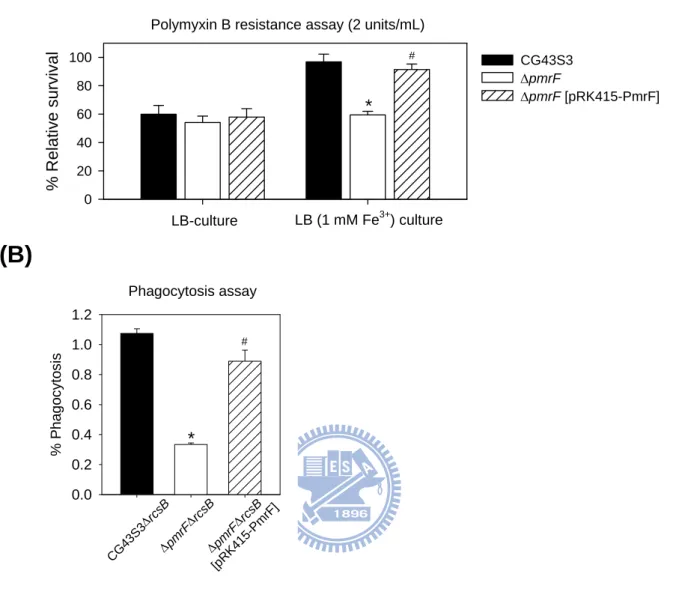
To investigate if the K. pneumoniae pmr homologues played a role in polymyxin B resistance, a pmrF deletion mutant strain and a plasmid pRK415-PmrF were generated. As shown in Fig. 2.2A, when the strains were grown in LB medium, a low magnesium condition (89), differences in the survival rates were not apparent. When the strains were grown in LB
supplemented with 1 mM FeCl3, an apparent deleting effect of pmrF in polymyxin B
resistance was observed, and the survival rate could be restored by the introduction of pRK415-PmrF. The results indicated a role of PmrF in the polymyxin B resistance in high iron condition. In addition to the mucosa surfaces, antimicrobial peptides and proteins play important roles in the microbicidal activity of phagosome (14). To investigate the effect of pmrF deletion in the bacterial survival within phagosome, phagocytosis assay was carried out. Since K. pneumoniae CG43S3 was highly resistant to engulfment by phagocytes in our initial experiments, the ΔrcsB mutant which produced less CPS was used as the parental strain to generate ΔpmrFΔrcsB mutant. As shown in Fig. 2.2B, deletion of pmrF resulted in an approximately four-fold reduction in the recovery rate, which was restored after the introduction of pRK415-PmrF. This indicated an important role of pmrF not only in polymyxin B resistance but also in bacterial survival within macrophage.
2.3.3 Deletion effect of Klebsiella pmrA, pmrD or phoP on polymyxin B resistance
To investigate how PmrA, PhoP and PmrD were involved in the regulation of polymyxin B resistance in K. pneumoniae, ΔpmrA, ΔphoP and ΔpmrD mutant strains were generated. Deletion of either one of these genes resulted in a dramatic reduction of resistance to polymyxin B when the strains were grown in LB medium (Fig. 2.3A). The deleting effects were no longer observed when the strains grown in LB supplemented with 10 mM magnesium, implying an involvement of the PhoP-dependent regulation in LB, a low magnesium environment. Under high-iron conditions, the deletion of pmrA caused the greatest reduction in the survival rate. Introduction of pRK415-PmrA or pRK415-PhoP into the ΔpmrAΔphoP double mutant strain not only restored but also enhanced the bacterial resistance to polymyxin B (Fig. 2.3B), which is likely due to an over-expression level of phoP or pmrA by the multicopy plasmid. Finally, whether the deletion of pmrA, phoP or pmrD affected the survival rate in phagosomes was also investigated. Interestingly, deletion of phoP resulted in most apparent effect while the pmrA deletion had less effect on the bacterial survival in macrophages. This was probably due to low iron concentration in the phagosomes (89). The introduction of pRK415-PhoP or pRK415-PmrD could restore the recovery rates of ΔphoPΔrcsB and ΔpmrDΔrcsB, although not to the extent displayed by the parental strain. Taken together, our results indicate the presence of two independent pathways in the regulation of polymyxin B resistance and the bacterial survival within macrophage phagosomes.
2.3.4 Effect of pmrA, phoP or pmrD deletion on PpmrH::lacZ and PpmrD::lacZ expression As the functional role of the structural gene pmrF and the regulator genes phoP, pmrD and pmrA was verified, it would be of importance to investigate the regulatory network govern by PhoPQ-PmrD-PmrAB on the expression of pmr genes. Sequence analysis has revealed PhoP and PmrA box consensus in the upstream region of pmrH and PhoP box consensus in the upstream region of pmrD (Fig. 2.4A). To investigate the interplay of PhoP, PmrA, and PmrD on the expression of pmr and pmrD genes, the reporter plasmids placZ15-PpmrH and placZ15-PpmrDwere constructed and mobilized into K. pneumoniae CG43S3ΔlacZ and its derived ΔpmrAΔlacZ, ΔpmrDΔlacZ or ΔphoPΔlacZ isogenic strains, respectively. The β-galactosidase activities of K. pneumoniae transformants under different environmental conditions were determined. In the wild-type strain CG43S3ΔlacZ, the PpmrH::lacZ activity was repressed in the presence of high magnesium but enhanced in high
ferric ion (Fig. 2.4B). Such iron-inducible activity was abolished after the addition of iron scavenger deferoxamine. As shown in Fig. 2.4B, deleting effect of pmrA or phoP on the activity of PpmrH::lacZ could be observed in LB or LB supplemented with ferric iron. The
negative effect of pmrD deletion was also apparent at high iron condition but was abolished after the addition of deferoxamine. The results clearly demonstrate the involvement of PmrA, PhoP and PmrD in the regulation of the expression of pmr genes, particularly in the presence of high ferric irons. As shown in Fig. 2.4C, the PpmrD::lacZ activity was significantly reduced
in high-magnesium conditions or upon the deletion of phoP. Interestingly, the deletion of pmrA or high ferric irons had little effect on the activity of PpmrD::lacZ. The results suggest
that the expression of K. pneumoniae pmrD is regulated in a PhoP-dependent but PmrA-independent manner.
2.3.5 Analysis of EMSA indicates a direct binding of the recombinant PhoP to PpmrD The binding of PhoP or PmrA to PpmrH has been determined recently (165). In order to
determine whether PhoP binds directly to PpmrD, EMSA was performed. As shown in Fig.
2.5A, binding of the recombinant His-PhoP protein to PpmrD was evident by the formation of a
protein/DNA complex with a slower mobility. The binding specificity was also examined by the addition of specific competitor (PpmrD DNA) or non-specific competitor (pmrD gene or
pUC19 DNA). The formation of the complex was gradually reduced with the increasing ratios of unlabeled to labeled PpmrD DNA fragments and completely diminished at the highest
protein/DNA complexes could also be observed although a portion of free probe was still noted, possibly due to the high amounts of DNA added. As shown in Fig. 2.5B, the formation of protein/DNA complex diminished when His-PhoPN149, in which the carboxyl-terminal
helix-turn-helix domain has been truncated, was used instead of His-PhoP. The results strongly suggest the PhoP binds via its C-terminal domain to the promoter of pmrD for the activation of the pmrD expression in K. pneumoniae.
2.3.6 Two-hybrid analysis of the in vivo interaction between Klebsiella PmrD and PmrA
The interaction between Klebsiella PmrD and PmrA has been shown as a prerequisite for the connector-mediated pathway (165). To demonstrate in vivo interaction, a bacterial two-hybrid assay was performed. The plasmid pBT-PmrA carrying the RNAPα-PmrA coding region and the plasmid pTRG-PmrD carrying the λ-cI-PmrD coding sequence were generated. In vivo interaction between the two reporter strains allowed the binding of λ-cI to the operator region as well as the recruitment of α-RNAP for the expression of the ampR and lacZ reporter genes. The bacteria harboring the positive control plasmids pTRG-Gal11P and pBT-LGF2 showed a vigorous growth on the indicator plate, as reflected by the apparent colony formation when the culture was diluted serially (Fig. 2.6A). In contrast, the strain carrying the negative control vectors pBT and pTRG revealed impaired colony formation. As shown in Fig. 2.6A, a profound growth pattern of the E. coli cells harboring pBT-PmrA and pTRG-PmrD was observed indicating an interaction between the PmrD and PmrA.
2.3.7 PmrD prevents the dephosphorylation of PmrA catalyzed by PmrB
In S. enterica, the phosphorylation of PmrA by the cognate sensor protein PmrB has been demonstrated to enhance its affinity in binding to its target promoter. The subsequent dephosphorylation of PmrA by PmrB helped to relieve from over-activation of this system (1). In Salmonella, PmrD has been shown to be able to protect PmrA from both intrinsic and PmrB-mediated dephosphorylation (22). To verify if Klebsiella PmrD also participates in the phosphorylation, in vitro phosphotransfer assay was carried out with the recombinant proteins His-PmrA, His-PmrD and His-PmrBC276. As shown in Fig. 2.6B, the His-PmrA was rapidly
phosphorylated upon addition of the autophosphorylated His-PmrBC276 and then gradually
dephosphorylated. Addition of His-PmrD apparently prolong the phosphorylation state of the His-PmrA, which could be maintained for at least 60 min (Fig. 2.6B). As shown in Fig. 2.6C, the specificity of the interaction between His-PmrD and His-PmrA was also demonstrated since the phosphorylation state of His-PmrA could not be detected when incubated with the
small cationic proteins RNase A or cytochrome C (117). Similar levels of phospho-PmrBC276
were observed in the presence or absence of PmrD (Fig. 2.6D), suggesting the His-PmrD had no effect on the phorphorylation state of His-PmrBC276. The above findings have indicated
that the reduced polymyxin B resistance in ΔpmrD mutant (Fig. 2.3A) could be due to the insufficient expression level of pmr genes (Fig. 2.4B) resulting from rapid dephosphorylation of PmrA by PmrB in the absence of PmrD. Accordingly, the PmrD connector-mediated pathway also played a role in K. pneumoniae resistance to polymyxin B.
2.3.8 Identification of residues critical for PmrD functionality
Since the PmrD homologues from K. pneumoniae, E. coli and S. enterica shared a lower sequence identity (30.3%) compared to the respective PmrA (74.9%) or PmrB (58.5%) counterparts, it was likely that differences in PmrD rather than PmrA sequences would alter their interaction properties and the subsequent polymyxin B resistance. To provide insights into how K. pneumoniae PmrD exert its function and to explore how PmrD sequence variation during the evolution could result in functional divergences, multiple sequence alignment of PmrD homologues was performed (Fig. 2.7A). Of all the amino acids aligned, the residues that may exhibit charge-charge interaction properties or conserved among the PmrD proteins were selected and individually changed to Alanine. The ability of the altered pmrD alleles encoding different PmrD proteins with point mutations to restore the polymyxin B resistance in ΔpmrD mutant was then investigated.
As shown in Fig. 2.7B, the polymyxin B resistance of ΔpmrD mutant strains carrying pRK415 or the derived plasmids encoding either the wild-type PmrD or PmrD with each point mutation was determined. Compared with the strain carrying a wild-type allele, the survival rates of ΔpmrD mutants carrying plasmids encoding PmrDK6A, PmrDS23A, PmrDE31A, PmrDG24A, PmrDL26A, PmrDM28A, PmrDT46A, PmrDY53A or PmrDY71A were reduced, indicative of an important role of these residues in PmrD functionality. In contrast, ΔpmrD strains carrying PmrDQ9A, PmrDD10A, PmrDT69A, PmrDT77A, PmrDT78A, PmrDD80A encoding plasmids were almost as resistant as the strain harboring pRK415-PmrD. The results suggested that some of the charged residues, such as Lysine at position 6 and Glutamate at position 31, may play an important role in the local charge-charge interactions with the charged domains on PmrA, such as the receiver domain with a negatively charged aspartic acid. In addition, some of the well-conserved residues may have maintained the overall stability of PmrD including Serine at position 23, Glycine at position 24, Leucine at position 26, Methionine at position 28, Threonine at position 46,
Tyrosine at positions 53 and 71. Whether the alteration of these residues would affect the turnover of PmrD protein remained to be verified. Since the residues involved PmrD functionality have been identified, it would be important to explore their roles in PmrD/PmrA interaction in order to elucidate the mode of action between these proteins.
In addition to the residues selected above, it was found that PmrDC35S could protect PmrA from dephosphorylation as PmrD (Fig. 2.7C). The alteration in Cysteine at position 54 seemed to affect the overall stability of PmrD since the protein was mostly present at the insoluble fraction indicating the importance of disulfide bond formation in PmrD structure. Recently our collaborators have determined the solution structure of Klebsiella PmrD and conducted amide chemical shift perturbations as well as saturation transfer assays using PmrDC54S to reveal the recognition surface of PmrD by the dimerized receiver domain of PmrA. The interface contained a contiguous patch involving Tryptophan at positions 3 and 4, Serine at positions 23 and 73, Leucine at position 26, Glutamate at positions 27 and 74, Methionine at position 28, Threonine at positions 46 and 69, Leucine at position 48, Alanine at positions 49, 51 and 68, Aspartic acid at position 50, Arginine at position 52, Isoleucine at position 65, Asparagine at position 67, Histidine at position 70 and Tyrosine at position 71. The findings have provided a structural basis of our results in polymyxin B resistance assay shown in Fig. 2.7B and a rational explanation for the critical role of residues at positions 23, 26, 28, 46 and 71 in PmrD/PmrA recognition process.