(2, 3, 4, 5, 7, 8)
50% (2)
(20)
WTO
(polymerase chain reaction PCR) (17)
E. carotovora subsp.
carotovora (Ecc) E. chrysanthemi (Ech)(13)
Erwinia
Erwinia carotovora subsp. carotovora
Erwinia chrysanthemi
1 1 2 1, 3 1 2 3 cylin@wufeng.tari.gov.tw 96 2 27. 2007. Erwinia carotovora subsp. carotovora
Erwinia chrysanthemi . 16 19-29
(RAPD) Erwinia
carotovora subsp. carotovora (Ecc) DNA 1,200 bp Y20
pY20Ecc-1200 bp NCBI
E. chrysanthemi (Ech) rhi N 83 %
Ec3F / Ec4R Ecc Ech
225 Ecc 497 bp 85 Ech
548 bp
Ecc Ech DNA 10~50 pg 6 7
DNA Ec3F / Ec4R Ecc Ech
1.2 101
1.1 101
2 3
Ec3F / Ec4R Ecc Ech
Ecc Ech
E. carotovora subsp. atroseptica Ech
ERWFOR (23)
E. carotovora
subsp. atroseptica ECA1f / ECA2r(11) Ech ADE1/ADE2(18)
5A / 5B(1) Ecc Ech
(random amplified polymorphic DNA, RAPD) Ecc
Ecc Ech
E. carotovora subsp. carotovora
(Ecc) (153 ) (5 )
(5 ) (14 ) (25
) (20 ) (3 )
225 Ecc E. chrysanthemi (Ech) (15
) (70 ) 85 Ech DNA 5 ml LB broth (Luria-Bertaini broth) 30 16 hr 12,000 rpm 10 min 1 ml STE 12,000 rpm 10 min 0.5 ml
STE 50 l 10% SDS (Sodium dodecyl
sulfate) 65 30 min 25 l / ml
proteinase K (100 g / ml) 30 min 27.5 l / ml RNase A (50 g / ml) 37
30 min phenol / chloroform
(phenol:chloroform:isoamylalcohol 25 24 1) 12,000 rpm 4 30 min 1 / 10 3 M NaOAc 2 95% -20 1 hr DNA 70% 12,000 rpm 4 30 min 50 15 min 100 l 4 DNA OD260 / 280 -30 (21) DNA
DNA Plasmid miniprep
purification (Genemark Technology Co., Ltd) Kanamycin (50 g / ml) 5 ml LB 37 16 hr 2 ml 12,000 rpm 5 min 200 l solution I 200 l solution II 200 l solution III 12,000 rpm 4 10 min DNA 700 l washing solution 12,000 rpm 4 30 sec 12,000 rpm 4 3 min DNA 1.5 ml 60 5~10 min 50 l 12,000 rpm 4 30 sec DNA RAPD
Ecc (Zan01, Zan03, Zan13) Ech 3 (Ech15, Ech20, Sr54)
DNA OPERON 10-MER KITS OPX
01~20, OPY 01~20 OPZ 01~20 60
RAPD 20 l 100 ng
DNA 200 M dNTPs 0.25 M 0.8 U
DNA GeneAmp PCR
System 2400 94 1 min
94 1 min, 40 1 min, 72 2 min 40
72 20 min 8 l
1X TAE 1.5% agarose gel 100 V
(ethidium
bromide) UV box
Ecc Ech Table 1. Bacterial strains used in this study
Bacterium Strain
Agrobacterium tumenfaciens AA19
Burkholderia caryophylli Tw7, Tw9
Erwinia carotovora subsp.
Zan01~50
carotovora
Erwinia chrysanthemi Ech10~22, Sr53~56
Pantoea agglomerans Yx5, Yx7
Pseudomonas aeruginosa Pae
P. putida Pu
P. syringae pv. syringae Pss
Ralstonia solanacerarum Ia29, 57
Xanthomonas axonopodis pv.
A072
DNA RAPD DNA DNA DNA Ecc RAPD DNA Ultrafree® -DA (MILLIPORE USA) KIT Gel Nebulizer
Ultrafree-MC Vial 6,000 rpm
10 min DNA
Random Primer Fluorescein Labeling Kit with
Antifluorescein-AP (NEN) 19 l
DNA 5 min
5 min 5 l Random primer and reaction buffer mix 5 l Fluorescein nucleotide mix 1
l Klenow fragment 30 l 37 1 hr 5 l 0.1 M EDTA (pH8.0) -30 (Southern hybridization) Southern (24) DNA
Ecc Ech DNA
OPY-20 RAPD
0.25 N HCl 10 min
denature buffer (0.5 M NaOH 1.5 M NaCl)
30 min neutralization buffer
(0.1 M Tris-HCl, pH7.5; 1.5 M NaCl) 30 min 10X SSC 10X SSC 16 hr 254 nm 2X SSC Salmon Sperm DNA 200 l (hybridization solution: 5X SSC, 0.5% SDS, 50% Formamide, 5X
Denhardts) 3 min 5 min
hybridization solution 65
1 hr 0.1% SDS
2X SSC 5 min 0.1%
SDS 0.2X SSC 65 15 min
buffer I (0.1 M Tris-HCl , pH7.5 ; 0.15 M NaCl) 5 min buffer (0.5% blocking reagent in buffer
II, 4 ) 1 hr Anti-fluorescein-AP
conjugate (NEN®
Life Science Products, Boston)
bufferII (1:5000) 1 hr buffer I
5 min buffer (0.1 M Tris-HCl , pH9.5;
0.15 M NaCl) 5 min
CDP-Star 12.5 mM (Tropix, U.S.A.) buffer (1:200)
5 min X-ray 10 min
DNA
TOPO TA Cloning®
(Invitrogen) DNA
4 l DNA 1 l Salt
solution 1 l pCR®
II-TOPO vector 5 min
2~6 l (competent cell) 30 min 42 30 sec 2 min 250 l SOC 200 rpm 37 1 hr 40 l 100 mM IPTG 40 l 40 g / ml X-gal Kanamycin (50 ppm) LA 37 DNA EcoRI National Center for Biotechnology Information (NCBI)
(GenBank)
Vector-NTI 8 (InforMax) G+C hairpin duplex structure (forward primer) (reverse primer)
100 ng
DNA 0.1 M 200 M dNTPs 0.4 U
Pro-Taq DNA polymerase 1X
20 l PCR 94
/ 5 min 94 / 30 sec 60 / 30 sec 72 / 30 sec
30 72 10 min
1X TAE 2% agarose
(1) Ec3F / Ec4R
DNA 225 Ecc
85 Ech DNA PCR
Ec3F / Ec4R Ecc Ech DNA
DNA Agrobacterium tumefaciens, Burkholderia caryophylli, Pantoea agglomerans, Pseudomonas aeruginosa, P. putida, Ralstonia solanacearum,
Xanthomonas axonopodis pv. dieffenbachiae 6 7
DNA
10 ng PCR
Ec3F / Ec4R Ecc Ech DNA
Ecc Ech
NA 24 hr
106
cfu / ml P. syringae pv. syringae P.
putida A. tumefaciens 104 ~108 cfu / ml Ecc Ech 106 cfu / ml 50 l 0.5 N NaOH PCR Ec3F / Ec4R (2) Ec3F / Ec4R
DNA Ecc Ech
DNA 100, 50, 10, 1 ng / l 100, 50, 10, 5, 1,
0.1 pg / l 1 l Ec3F / Ec4R
PCR Ec3F / Ec4R Ecc
Ech DNA Ecc Ech NA 24 hr OD620 0.3 ( 108 cfu / ml) 10 101 ~107 cfu / ml
spiral plater (model D, Spiral systems, Inc. Bethesda,
Maryland) NA 30 24 hr 50 l 200 l 50 l 0.5 N NaOH 50 l 50 l 1M Tris-HCl (pH 8.0) 2 l Ec3F / Ec4R PCR Wang(27) Ecc Ech 50 l 0.5 N NaOH 25 l 25 l 1 M Tris buffer 1 l Ec3F / Ec4R PCR PCR 20 20 50 mg 100 l 0.5 N NaOH 5,000 rpm 3 min 15 l 135 l 100 mM Tris-HCl (pH 8.0) (21) 2 l PCR Ec3F / Ec4R RAPD Ecc-1200
Ecc (Zan01, Zan03, Zan13) Ech 3 (Ech15, Ech20, Sr54)
DNA OPX Y Z 01~20 60
RAPD OPY-20 Ecc
1,200 bp ( A) 1,200 bp DNA 1,200 bp Ecc ( B) 1,200 bp Ecc DNA pCR® II-TOPO pY20Ecc-1,200 bp DNA NCBI
Ech rhiN gene
83% ( )
339 427 bp 578 1,171 bp Ech Ecc 49 bp
Vector NTI 8 (InforMax) 2
Ec3F Ec4F 2 Ec3R
Ec4R ( ) PCR
(a) 94 5 min 1 (b) 94 1 min 60 30 sec 72 30 sec 30 (C) 72 10 min 1
Ec3F / Ec4R
( ) Ec3F / Ec4R
PCR Ecc Ech
Ec3F / Ec4R 153 5 5 14 25 20 3 225 Ecc Ec3F / Ec4R 497 bp 15 70 85 Ech 548 bp Ec3F /
Ec4R Ecc Ech
Ecc (Zan01) Ech (Ech15)
DNA A. tumefaciens,
B. caryophylli, P. agglomerans, P. aeruginosa, P. putida,
R. solanacearum X. axonopodis pv. dieffenbachiae
6 7 DNA PCR
Ec3F / Ec4R Ecc Ech
( )
Ecc Ech 106
cfu / ml 100 100 P.
syringae pv. syringae, P. putida, A. tumefaciens 2
3 (104 ~108 cfu / ml) PCR Ecc 497 bp Ech 548 bp 2 3 ( ) Ec3F / Ec4R
Ec3F / Ec4R Zan01 (Ecc)
Ech15 (Ech) DNA PCR
Ecc Ech DNA 10 pg~50 pg ( ) NaOH DNA PCR Ecc 1.1 101 Ech 1.2 101 ( )
Ecc Ech 0.5 N NaOH
DNA Ec3F / Ec4R PCR
Ecc Ech 497
bp 548 bp
E. carotovora
subsp. carotovora E. chrysanthemi ( )
PCR Ec3F / Ec4R PCR 497 bp Ecc Ec3F / Ec4R PCR 548 bp Ech ( ) Erwinia ( 13, 26)
Erwinia carotovora subsp.
carotovora (Ecc) E. chrysanthemi (Ech) Ecc
OPY-20
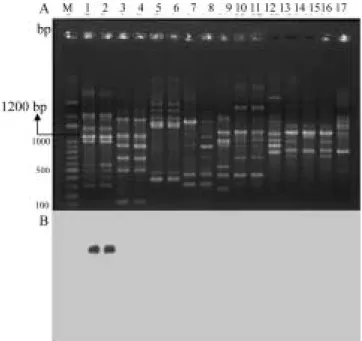
RAPD (A)
Ecc-Zan01 Y20Ecc-1200 (B) M Bio 100 marker 1-2 Ecc Zan01 Zan03 3-4 Ech Ech15
Ech20 5~17 ( )
Fig. 1. Agarose gel electrophoresis analysis showing RAPD patterns of strains of Erwinia carotovora subsp.
carotovora (Ecc), E. chrysanthemi (Ech) and other
bacteria using OPY-20 random primer (A) and southern hybridization of the RAPD products probed with Y20Ecc-1200 fragment cloned from Ecc strain Zan01. (B). Lanes 1-2, Ecc strains Zan01, Zan03; Lanes 3-4, Ech strains Ech15, Ech20; Lanes 5~17, non-target bacterial strains (Table 1); M, Bio 100 marker.
pY20Ecc-1200bp DNA Ecc Ech Ec3F
Ec4F Ec3R Ec4R
Fig. 2. The sequence of the inserted DNA fragment in recombinant plasmid pY20Ecc-1200bp. The sequences of primers Ec3F, Ec4F, Ec3R and Ec4R are underlined, and the similar sequence between Ecc and Ech are bold-faced.
(2, 3, 4, 5, 7, 8) Ech (1, 3) Ecc Ech CVP Erwinia (22) Ecc Ech CVP Ecc Ech Ecc Ech Erwinia (15) RFLP Erwinia (9, 12, 25, 28) DNA PCR Erwinia 23S rRNA E. amylovora(16)
pel Ec1 / Ec2
E. carotovora(12)
E. carotovora subsp. atroseptica (Eca) Ech(11, 23)
E. carotovora subsp. carotovora Zan01
E. chrysanthemi Ech15 6 7
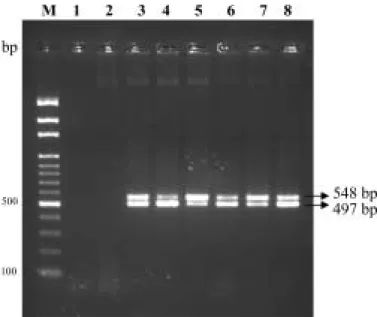
DNA Ec3F / Ec4R
M 100 bp marker 1
2 Pseudomonas putida 3-8, Ecc Ech
Agrobacterium tumefaciens, Burkholderia caryophylli, P. aeruginosa, P. putida, Ralstonia
solanacearum, Xanthomonas axonopodis pv.
dieffenbachiae
Fig. 3. Polymerase chain reaction (PCR) products amplified from total DNA of strains E. carotovora subsp.
carotovora and E. chrysanthemi mixed with other
nontarget bacterial DNA with primer pair Ec3F/Ec4R. M, 100 bp marker; lane 1, negative control; lane 2, only P.
putida DNA; lanes 3-8, Ecc and Ech mixed with A. tumefaciens, B. caryophylli, P. aeruginosa, P. putida, R. solanacearum, X. axonopodis pv. dieffenbachiae,
respectively.
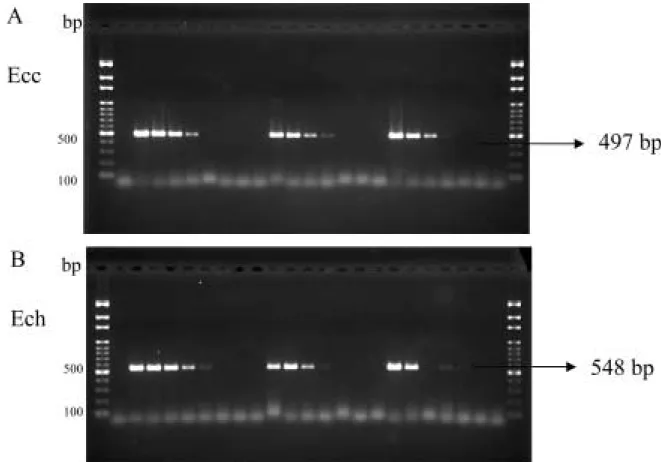
Ec3F / Ec4R E. carotovora subsp. carotovora Zan01 E.
chrysanthemi Ech15 DNA A Ecc (Zan01) DNA B Ech (Ech15) DNA
M 100 bp marker 1-9 100 ng, 50 ng, 10 ng, 1 ng, 100 pg, 50 pg, 10 pg, 1 pg 0.1 pg Fig. 4. Sensitivity of PCR using primer pair Ec3F/Ec4R to detect total DNA of E. carotovora subsp. carotovora Zan01 (A) and Erwinia chrysanthemi Ech15 (B). Lanes 1-9, 100 ng, 50 ng, 10 ng, 1 ng, 100 pg, 50 pg, 10 pg, 1 pg and 0.1 pg respectively. M, 100 bp marker.
Ec3F / Ec4R Ecc Ech 2, 10, 17 1.1~1.2 104 cfu 3, 11, 18 1.1~1.2 103 cfu 4, 12, 19 1.1~1.2 102 cfu 5, 13, 20 1.1~1.2 101 cfu 6, 14, 21 1.1~1.2 cfu 7, 15, 22 1.1~1.2 10-1 cfu 8, 16, 23 1.1~1.2 10-2 cfu M 100 bp marker 1
Fig. 5. Sensitivity of PCR for detection of cultured E. carotovora subsp. carotovora (A) and E. chrysanthemi (B) with primer pair Ec3F / Ec4R. Lanes 2, 10 and 17, 1.1~1.2 104
cfu; lanes 3, 11 and 18, 1.1~1.2 103
cfu; lanes 4, 12 and 19, 1.1~1.2 102
cfu; lanes 5, 13 and 20, 1.1~1.2 101
cfu; lanes 6, 14 and 21, 1.1~1.2 cfu; lanes 7, 15 and 22, 1.1~1.2 10-1 cfu; lanes 8, 16 and 23, 1.1~1.2 10-2
cfu; . M, 100 bp marker; lane 1, negative control.
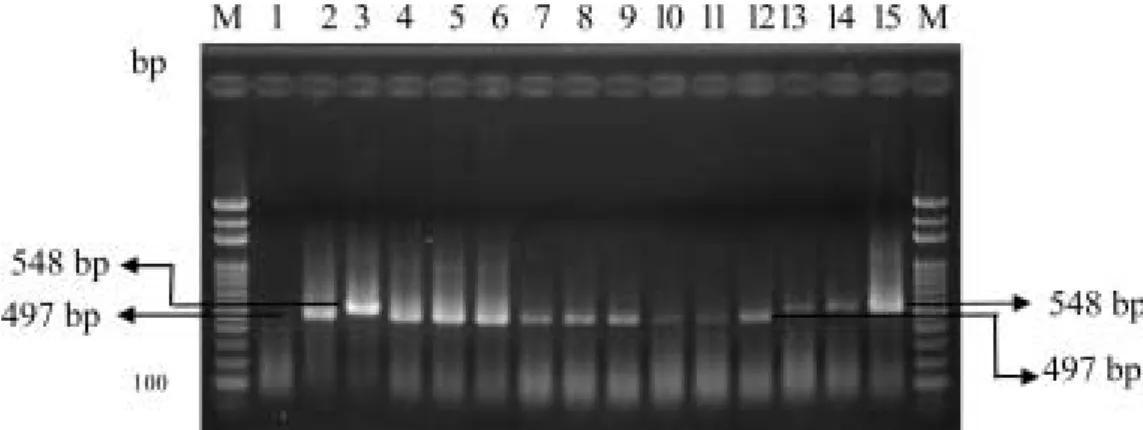
Ec3F / Ec4R Ecc Ech M 100 bp Marker 1 ddH2O 2 Sr50 DNA 3-13 Ecc Zan 1 2 3 9 1 0 1 3 3 1 34 36 39 50 14 - 17 Ech Ech 17, 18, 19, 20
Fig. 6. Identification of Erwinia carotovora subsp. carotovora and Erwinia chrysanthemi by PCR with primer pair Ec3F / Ec4R. Individual colonies on agar plates were extrated with NaOH and used as templates for PCR. Lane 1, ddH2O negative control; lane 2, Sr50 DNA; lanes 3-13, colonies from Ecc strains Zan1, 2, 3, 9, 10, 13, 31, 34, 36, 39, 50 respectively; lanes 14 - 17, colonies from Ech strains Ech 17, 18, 19 and 20 respectively. M, 100 bp marker.
16S rRNA Eca Ecc Ech(25)
Erwinia Ech ADE1 / ADE2(18) 5A / 5B(1) E. carotovora Ec1 / Ec2(12) Ecc Ech PCR Xanthomonas axonopodis pv.
vesicatoria Xanthomonas vesicatoria
(14) RAPD Ecc 1,200 bp Ech rhiN 83% Ech Ecc 49 b p
Ec3F / Ec4R PCR Ecc
497 bp Ech 548 bp PCR Ecc Ech Ec3F / Ec4R PCR 10 pg~50 pg DNA (10) Ecc 11 Ech 12 (6) (6) 5A
/ 5B Ec1 / Ec2 Ecc
1.3 102 Ech 5.2 101 Toth (25) Toth (25) 2.0 103 ~ 3.4 104 Ec3F / Ec4R Ecc Ech
Ec3F / Ec4R PCR ~ Ecc Ech Ecc Ech Ec3F / Ec4R (19) 1. 1995 Erwinia chrysanthemi PCR 2. 1994
Ec3F / Ec4R Ecc Ech
M 100 bp Marker 1 ddH2O 2 Zan01 DNA 3
Ech15 DNA 4-6 7-9 10-12
13-15
Fig. 7. Detection of infected tissues of cala lily and phalaenopsis from fields by PCR with primer pair Ec3F / Ec4R. Lane 1, ddH2O negative control; lane 2, Zan01 DNA; lane 3, Ech15 DNA; lanes 4-6, infected rhizome tissues of cala lily; lanes 7-9, infected stem tissues of cala lily; lanes 10-12, infected leaf tissues of cala lily, lanes 13-15, infected leaf tissues of phalaenopsis; M, 100 bp marker.
3. 2004
46 367-378
4. 2 0 0 3 E r w i n i a
carotovora subsp. carotovora
45 257-262 5. 2001 43 105-115 6. 1998 Erwinia 7. 1997 PCR Erwinia chysanthemi 8. 1983
9. Anna, O. A., Hyman, L. J., Toth, R. L. and Toth, I. K. 2002. Application of amplified fragment length polymorphism fingerprinting for taxonomy and identification of the soft rot bacteria Erwinia
carotovora and Erwinia chysanthemi. Appl. Environ.
Microbiol. 68 : 1499-1508.
10. Berswell, S., Pahl, A., Bellemann, P., Zeller, W. and Geider, K. 1992. Sensitive and species-specific detection of Erwinia amylovora by polmerase chain detection analysis. Appl. Environ. Microbiol. 58 : 3522-3526.
11. De Bore, S. H. and Ward, L. J. 1995. PCR detection of
Erwinia carotovora subsp. atroseptica associated with
potato tissue. Phytopathol. 85 : 854-858.
12. Darrasse, A., Prious, S., Kotoujansky, A. and Bertheau, Y. 1994. PCR and restriction fragment length polymorphism of a pel gene as a tool to identify
Erwinia carotovora in relation to potato disease. Appl.
Environ. Microbiol. 60 : 1437-1443.
13. Hsu, S. T. and Tzeng, K. C. 1981. Species of Erwinia
associated with soft rot disease of plant in Taiwan.
Pages 9-18 in Proc. Fifth Int. Conf. Plants Path. Bact. J. C. Lozano, ed. CIAT. Cali, Colombia.
14. Kulfu, K. M. and Cuppels, D. A. 1997. Development of a diagnostic DNA probe for Xanthomonas causing bacterial spot of peppers and tomatoes. Appl. Environ. Microbiol. 63 : 4462-4470.
15. Louws, F. J., Rademarker, J. L. W. and de Bruijn, F. J. 1999. The three Ds of PCR-based genomic analysis of phytobacteria: diversity, detection, and disease diagnosis. Annu. Rev. Phytopathol. 37 : 81-125. 16. Maes, M., Garbeva, P. and Crepel, C. 1996.
Identification and sensitive endophytic detection of
fireblight pathogen Erwinia amylovora with 23S ribosomal DNA sequences and polymerase chain reaction. Plant Pathol. 45 : 1139-1149.
17. Miller, S. A. and Martin, R. R. 1988. Molecular diagnosis of plant disease. Annu. Rev. Phytopathol. 26 : 409-432.
18. Nassar, A., Darrasse, A., Lemattre, M., Kotoujansky, A. and Dervin, C., 1996. Characterization of Erwinia
chrysanthemi by pectinolytic isozyme polymorphism
and restriction fragment length polymorphism analysis of PCR amplified fragments of pel genes. Appl. Environ. Microbiol. 62 : 2228-2235.
19. Perombelon, M. C. M. and Apple, J. D. 1984. Soil and seed tubers as source of inoculum of Erwinia
carotovora pv. carotovora for stem soft rot of potatoes.
Phytopathology 74 : 429-432.
20. Perombelon, M. C. M. and Kelman, A. 1980. Ecology of the soft rot Erwinias. Annu. Rev. Phytopathol. 18 : 361-387.
21. Sambrook, j., Mantis, T. I. and Fritsch, E. F. 1989. Molecular cloning: a laboratory manual. 2nd
ed. Cold Spring Harbor Laboratory, Press, N. Y.
22. Schaad, N. W., Azad, H., Peet, R. C. and Panopoulos, N. J. 1989. Identification of Pseudomonas syringae pv.
phaseolicola by a DNA hybridization probe.
Phytopathology 79 : 903-907.
23. Smid, E. J., Jansen, A. H. J. and Gorria, L. G. M. 1995. Detection of Erwinia carotovora subsp.
atrosepticai and Erwinia chrysanthemi in potato tubers
using polymerase chain reaction. Plant Pathol. 44 : 1058-1069.
24. Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98 : 503.
25. Toth, I. K., Hyman, L. J. and Wood, J. R. 1999. A one-step PCR-based method for the detection of economically important soft rot Erwinia species on micropropagated potato plants. J. Appl. Microbiol. 87 : 158-166.
26. Tzeng, K. C. and Hsu, S. T. 1981. Identification and characterization of soft-rotting Erwinia in Taiwan. Plant Prot. Bull. 23 : 77-85.
27. Wang, H., Qi., M. and Cutler, A. J. 1993. A simple method of preparing plants samples for PCR. Nucleic Acids Res. 21 : 4153-4154.
28. Williams, J. G. K., Kubelik, A. R., Livak, K. J., Rafalski, J. A. and Tingey, S. V. 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 18 : 6531-6535.
ABSTRACT
Hseu, S. H.,1
Shentue, H1
., Tzeng, K. C2
. and Lin, C. Y.1, 3
2007. Development of specific primers for differential identification pathogen Erwinia carotovora subsp. caratovora and Erwinia chrysanthemi. Plant Pathol. Bull. 16 19-29 (1
Plant Pathology Division, Agricultural Research Institute, Council of Agriculture, Wufeng, Taichung, Taiwan; 2
Department of Plant pathology, Chung-Hsing University, Taichung, Taiwan, 3
Corresponding author, E-mail: cylin@wufeng.tari.gov.tw )
A specific PCR (polymerase chain reaction) primer pair has been developed using RAPD (random amplified polymorphic DNA) to differentially detect Erwinia carotovora subsp. carotovora (Ecc) and E. chrysanthemi (Ech). A specific 1,200 bp DNA fragment was amplified and cloned from Ecc DNA using OPY20 primer, one of the sixty random primers used in RAPD. Nucleotide sequence of this 1,200 bp DNA showed 83% similarity to Ech rhiN gene, where conserved sequences were displayed in two regions. But in Ech rhiN sequence, there are additional 49 bp nucleotides inserted between these two conserved regions. Herein, a set of primers Ec3F / Ec4R was designed and applied to differentially detect Ecc and Ech by PCR. Using bacterial chromosomal DNA as templates, a distinct 497 bp fragment can be amplified from 225 Ecc strains, and a 548 bp fragment was amplified from 85 Ech strains. No fragment can be amplified from the other seven bacterial species in six genera. Sensitivity of PCR for detecting Ecc and Ech was between 10 50 pg for purified DNA and 1.1 101
cfu and 1.2 101
cfu for cultured cells, respectively. It took 3-4 hr to detect Ecc and Ech by colony PCR using Ec3F / Ec4R primer set and the target bacteria in infected tissues of cala lily and phalaenopsis. Results presented here suggest that the primer pair Ec3F / Ec4R applied in PCR can be very useful in rapid identification, detection, and differentiation of the soft rot pathogens Ecc and Ech. Key words: Cala lily, Erwinia carotovora subsp. carotovora, Erwinia chrysanthemi, primer.