行政院國家科學委員會專題研究計畫 成果報告
臺灣 methicillin 抗藥性金黃色葡萄球菌抗藥性基因之分型
研究(2/2)
計畫類別: 個別型計畫 計畫編號: NSC93-2314-B-002-035- 執行期間: 93 年 08 月 01 日至 94 年 07 月 31 日 執行單位: 國立臺灣大學醫學院內科 計畫主持人: 張上淳 共同主持人: 王振泰 計畫參與人員: 吳佳玲、陳雨薇 報告類型: 完整報告 處理方式: 本計畫可公開查詢中 華 民 國 94 年 10 月 28 日
中文摘要
抗藥性金黃色葡萄球菌(MRSA)自 1990 年代以來,已成為台灣地區的重要 致病菌之一。其抗藥性的機轉在於攜帶了一段抗藥性基因──葡萄球菌卡匣染色 體(Staphylococcal cassette chromosome,簡稱 SCCmec 元素)。根據最近的研究, SCCmec 元素可分為四種型式:type I、II、III、IV。而截至目前為止,台灣地區 並沒有大規模針對本土的 MRSA 之 SCCmec 元素進行的分型研究。在我們先前 利用脈衝式磁場電泳(pulsed-field gel electrophoresis,PFGE)對全台灣院內感染 的 MRSA 菌株的分子分型研究中,我們發現了全台灣大部分的院內 MRSA 菌株 都是屬於 PFGE type C,並且全台灣有七個比較重要的 PFGE types。因此,我們 由此七個 PFGE type 中(type B、C、D、G、H、J、L)的每一個亞型(共計 31 個亞型)挑出一個 MRSA 菌株,利用聚合酶鏈鎖反應(polymerase chain reaction) 的方法來進行 SCCmec 元素的分型研究。另外,我們也收集 2003 年 1 月至 2003 年 12 月台大醫院所有符合社區性 MRSA 感染定義患者的 MRSA 菌株
(CA-MRSA),共計 19 株,進行其 SCCmec 元素的分型研究。研究結果顯示, 在台灣七種重要的院內 MRSA strains 中,PFGE type B、C、J、L 的菌株均攜帶 著第三型的 SCCmec 元素;而 PFGE type D、G、H 的菌株則攜帶著第四型的 SCCmec 元素。至於 19 株的 CA-MRSA 中,有 5 株屬 PFGE type C,有 8 株屬於 type D,有 4 株屬於 type H,而有兩株屬於 type S。這 19 株 CA-MRSA 中,除了 屬於 type C 的 5 株攜帶著第三型的 SCCmec 元素外,其餘均攜帶著第四型的 SCCmec 元素。根據上述的研究結果,我們歸結了以下幾個重點:全台灣最主要 的院內 MRSA strain,攜帶著第三型的 SCCmec 元素,而全台灣絕大部分的院內 MRSA 菌株不是攜帶著第三型的 SCCmec 元素就是第四型的 SCCmec 元素;台灣 地區的院內 MRSA 菌株和 CA-MRSA 可能已發生交叉傳播的現象;台灣地區的 CA-MRSA 大部分均攜帶著第四型的 SCCmec 元素;在所謂的 CA-MRSA 中,也 存在著一個主要的 MRSA strain。
英文摘要
Methicillin-resistant Staphylococcus aureus (MRSA) had caused a great impact on clinical medicine since mid-1990s in Taiwan. The mechanism of resistance to
methicillin in MRSA was due to the acquisition of additional genetics called staphylococcal cassette chromosome (SCCmec element). According to the recent studies, the SCCmec elements could be classifiec into four types: type I, II, III, and IV. Till now, there is still no published report about the detailed genetics of methicillin resistance, the typing of SCCmec element, of MRSA isolates in Taiwan. In our previous study, we found that there was a predominant MRSA strain, pulsed-field gel electrophoresis (PFGE) type C, among all the nosocomial MRSA isolates in Taiwan. And the majority of nosocomial MRSA isolates belonged to seven PFGE types, type B (3 subtypes), C (10 subtypes), D (2 subtypes), G (5 subtypes), H (5 subtypes), J (4 subtypes), and L (2 subtypes). Therefore, we selected one MRSA isolate from each PFGE subtypes, 31 isolates in total, to undergo the typing of their SCCmec elements. In addition, 19 community-acquired MRSA isolated collected from January 2003 to December 2003 at National Taiwan University Hospital were also included in this study. The study result demonstrated that among the nosocomial MRSA isolates, those belonging to PFGE type B, C, J, and L carried type III SCCmec element and those belonging to PFGE type D, G, and H carried type IV SCCmec element. Among the 19 CA-MRSA isolates, all except the 5 isolates belonging to PFGE type C carried type IV SCCmec element. In conclusion, our study found that the predominant nosocomial MRSA strain, PFGE type C, carried type III SCCmec element, the majority of
nosocomial MRSA isolates carried either type III or IV SCCmec element, the nosocomial MRSA isolates and CA-MRSA might had been cross-transmitted each other, most CA-MRSA carried type IV SCCmec element, and there was also a predominant strain among the CA-MRSA.
Key words: methicillin-resistant Staphylococcus aureus, MRSA, resistant gene, staphylococcal cassette chromosome
目錄
中文摘要 I 英文摘要 II 目錄 III 報告內容 1 研究計畫之目的、前言、文獻探討 1 研究方法 3 結果 6 討論 7 參考文獻 9 表一 12 表二 13 表三 14 圖 15 研究成果自評 16報告內容:
研究計畫之前言、目的、文獻探討
The first report of MRSA in the world is by Dr. Jevons in 1961 (1). And the first isolate of MRSA in Taiwan is found in 1981 (2). Thereafter, the rates of
nosocomial MRSA infections increased rapidly in Taiwan and most hospitals in Taiwan now have a high incidence of nosocomial MRSA infections (3, 4). The rates of MRSA over all nosocomial isolates of S. aureus in some Taiwan hospitals has already exceeded 80% in 1998 (5), which was much higher than that reported by the National Nosocomial Infection Surveillance System (NNIS) (6). Our previous studies have proved that the reasons leading to rapid increased of MRSA infection in Taiwan include overuse of antibiotics, poor adherence to isolation precaution of health care worker, and introduction of endemic strain (7-10). MRSA thus has become a major pathogen in Taiwan. In addition, because of its resistance, limited choice of drug to treat MRSA infection is another important clinical problem. However, there is still no detailed study on the genetic mechanism of methicillin resistance of MRSA in Taiwan. Understanding the detailed genetics of methicillin resistance of MRSA may be helpful to overcome this resistance in the future.
The genetic coding for methicillin resistance in S. aureus has been proven to be mecA gene (11). The expression of mecA gene results in a specific
penicillin-binding protein, PBP2’, that has a decreased binding affinity to β-lactam antibiotics and thus leading to methicillin resistance. The expression of mecA gene is regulated by two adjacent regulatory gene, mecI and mecR1. The mecA, mecI, and
mecR1 genes consist of the mecA gene complex (12). The mecA gene complex is
widely distributed among S. aureus species as well as among other staphylococcal species collectively called coagulase-negative staphylococci (13-15). Therefore, it has been speculated that mec may be freely transmissible among staphylococcal species. In the 1980s, direct chromosome analysis of MRSA strains revealed that a substantial length of the chromosomal DNA segment (greater than 30 kb) carrying
mec has no allelic equivalence in methicillin-susceptible S. aureus (MSSA) strains;
the segment was called “additional DNA” or “mec DNA” (16, 17). The size, structure, and biological properties of mec DNA had long remained unclear (18).
The most striking findings in recent studies on MRSA are the existence of SCCmec gene element (19). According to the studies of Hiramatsu et al, the “mec DNA” is now formally renamed as “SCCmec (Staphylococcal cassette chromosome
mec) element” and the SCCmec element is almost universally found in all MRSA
(cassette chromosome recombinase), ccrA and ccrB, genes and mecA gene complex (19, 20). The mecA gene complex can be classified into four types, type A, B, C, D. The ccr genes can be classified into to three types. The function of ccr genes is to precisely excise and insert the SCCmec element in way of both site and orientation specifically (19). Based on the structures and combinations of ccr genes and mecA gene complex, the SCCmec gene element in MRSA can be classified into four types (20, 21). Type III SCCmec gene carries more drug-resistant determinants than any other types. According to the study, conducted by Ito et al, on the analysis of SCCmec element of 38 major hospital-acquired MRSA strains isolated worldwide, SCCmec elements of MRSA isolates from Europe belong to type I and III, those from northern America belong to type II, most of those from Japan belong to type II, those from Australia and southeastern Asia belong to type III, and those from south Africa belong to type I (20). Type IV SCCmec element is so far only found in community-acquired MRSA isolates (21).
As mentioned above, there is still no published report about the detailed genetics of methicillin resistance of MRSA isolates in Taiwan. The type of SCCmec elements of MRSA isolated in Taiwan is also obscure. Our study is designed to illuminate the mecA complex (in the first year), and ccr genes (in the second year) of MRSA isolates in Taiwan as well as to determine the types of SCCmec elements in Taiwan and compare these results with MRSA isolates in other country. In addition, there are some sporadic cases of community-acquired MRSA infections. By typing the SCCmec gene element of MRSA isolates isolated from patients with nosocomial MRSA infections and those with community-acquired MRSA infections, whether the methicillin resistance comes from the same source between these isolates or not can also be determined.
研究方法
Definitions and MRSA isolates:
Patients with community-acquired MRSA (CA-MRSA) septicemia is defined as that patients had no history of hospitalization within prior 30 days develop signs and symptoms of sepsis before admission and their blood cultures taken within 48 hours after hospitalization yielded MRSA. Patients with nosocomial MRSA septicemia is defined as that patients developed signs and symptoms of sepsis three more days after admission and their blood cultures yielded MRSA. Based on our previous study, there were seven major types of nosocomial MRSA isolates determined by pulsed-field gel electrophoresis (PFGE) in Taiwan,
including type B (3 subtypes), C (10 subtypes), D (2 subtypes), G (5 subtypes), H (5 subtypes), J (4 subtypes), and L (2 subtypes) (10). Thirsty-one isolates, one isolate from every subtype, were selected for further microbiologic investigation. Nineteen CA-MRSA isolates collected from January 2003 to December 2003 at NTUH were also enrolled.
Determination of minimum inhibitory concentration (MIC):
All isolates were tested for their MIC levels of oxacillin, gentamicin, clindamycin, erythromycin, ofloxacin, levofloxacin, tetracycline, rifampin, trimethoprim/sulfamethoxazole, vancomycin, and linezolide using agar dilution method proposed by NCCLS (22).
PFGE:
All isolates were typed first by PFGE to determine whether those CA-MRSA belonged to the same molecular types of nosocomial isolates or not. The methods used for undergoing PFGE will be as those described in our previous study (9). The interpretation of PFGE patterns was according to the principals proposed previously (23, 24). Once a CA-MRSA is proved to belong to the same type as a nosocomial isolate, it was not used for further molecular study. All results will be double checked.
PCR and nuleotide sequencing for the analysis of mecA complex:
The chromosomal DNA will be prepared by means of the method
described by Hiramatsu et al and Matsuhashi et al (25, 26). PCR amplification was performed using 1 unit AmpliTaq (Perkin-Elmer Cetus, Foster City, Calif.) in 50 μl of reaction mixture (10mM Tris-HCl [pH 8.3], 50 mM KCl, 0.001% [wt/vol] gelatin, 50% [vol/vol] glycerol, 1.5 mM MgCl2, 200 mM each
deoxynucleoside triphosphatge, 1.0 mM each primer, and template DNA). The reaction was carried out by using a Gene Amp PCR system 9600
(Perkin-Elmer). Thermal cycling was set at 30 cycles (30 s for denaturation at 94℃, 1 min for annealing at 50℃, and 2 min for elongation at 72℃)
Long-range PCR amplification was performed using 2.6 U of Expand high-fidelity PCR system enzyme mix as recommended by the manufacturer (Boehringer Mannheim Biochemica, Mannheim, Germany). A 5-μl portion of the reaction volume was subjected to electrophoresis in a 0.8% agarose gel containing 1 μl of ethidium bromide per ml to detect the amplified DNA fragment. All PCR products were further sequenced using a 377 automated fluorescent DNA sequencing system (Perkin-Elmer, Foster City, Calif.) to compare the nucleotide homology with the published sequence in GenBank. The PCR and sequencing results were double-checked.
PCR primers and detection of mecA gene complex:
The primers used for the detection of mecA gene complex include: mA2 (5’-AACGTTGTAACCACCCCAAGA-3’), mA4 (5’-AGTGTATGATGAGCTATGAGA-3’), mA5 (5’- CGCTCAGGAAATTTGTTGTGC-3’), mA6 (5’-TATACCAAACCCGACAAC-3’), iS1 (5’-ACATTAGATATTTGGTTGCGT-3’), iS3 (5’-TCGGATGCTATCATTAAGCAT-3’), iS4 (5’-ACAATCTGTATTCTCAGGTCGT-3’), mI-1 (5’-AATGGCGAAAAAGCACAACA-3’), mI-2 (5’-GACTTGATTGTTTCCTCTGTT-3’), mcR2 (5’-CGCTCAGAAATTTGTTGTGC-3’), and mcR3 (5’-ATACTCCACGTTAATTCCATT-3’) (27).
Technical detection of the class A mecA gene complex was based on the positive PCR test results for both sets of primers, mI-1 plus mI-2 and mcR2 plus mcR3. The class B mecA gene complex was detected by long-range PCR using two sets of primers, iS-3 plus mA5 and iS-4 plus mA5. Detection of the Class C mecA gene complex was based on the negative PCR test results for two sets of primer, mI-1 plus mI-2 and mcR2 plus mcR3, as well as a positive long-range PCR test result for one set of primers, iS-1 plus mA6. Detection of the Class D mecA gene complex was based on the negative PCR test results for two sets of primer, mI-1 plus mI-2 and mcR2 plus mcR3, as well as a negative long-range PCR test result for one set of primers, iS-1 plus mA6 (27,
28).
PCR and nucleotide sequencing for the analysis of ccr genes:
The PCR and long-range PCR methods were used to analyze ccr genes are as described above. All PCR products were further sequenced using a 377 automated fluorescent DNA sequencing system (Perkin-Elmer, Foster City, Calif.) to compare the nucleotide homology with the published sequence in GenBank. The PCR and sequencing results were double-checked.
PCR primers and detection of ccr genes:
The primers used for the detection of ccr gene complex include: β2 (5’-ATTGCCTTGATAATAGCCITCT-3’),
α2 (5’-AACCTATATCATCAATCAGTACGT-3’), α3 (5’- TAAAGGCATCAATGCACAAACACT-3’), α4 (5’-AGCTCAAAAGCAAGCAATAGAAT-3’)
Technical detection of type 1 ccr complex was based on the positive long-range PCR test result for one set of primers, α2 plus β2. Technical
detection of type 2 ccr complex was based on the positive long-range PCR test result for one set of primers, α3 plus β2. Technical detection of type 3 ccr complex was based on the positive long-range PCR test result for one set of primers, α4 plus β2 (10, 27).
Determining the types of SCCmec elements:
After determining the types of mecA gene complex and ccr complex, the types of SCCmec elements was further determined using following criteria:
1. Type I SCCmec element: type 1 ccr complex + class B mecA gene complex;
2. Type II SCCmec element: type 2 ccr complex + class A mecA gene complex;
3. Type III SCCmec element: type 3 ccr complex + class A mecA gene complex;
4. Type IV SCCmec element: type 2 ccr complex + class B mecA gene complex;
結果
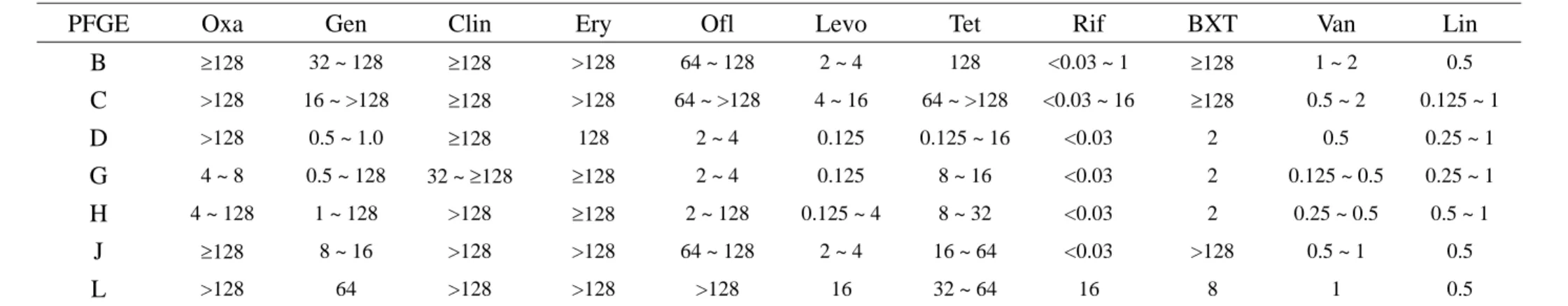
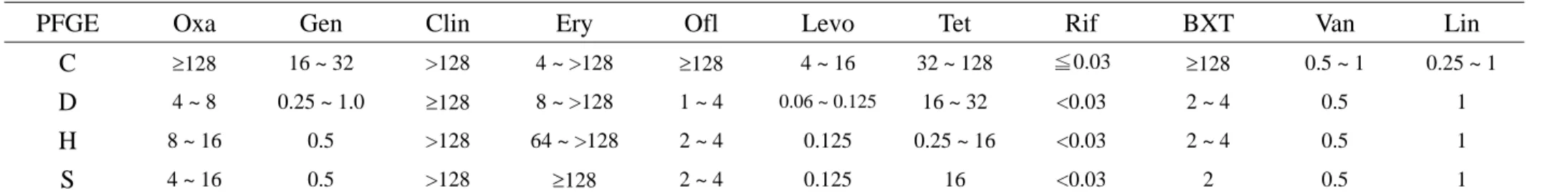
Drug susceptibility determined by MIC and PFGE types:
The drug susceptibility of the 50 MRSA isolates was listed in Table 1 and 2. Among the nosocomial MRSA isolates, both isolates of PFGE type L were only susceptible to vancomycin and linezolid but resistant to all other tested antibiotics. Most isolates of PFGE type C were only susceptible to vanocmycin, rifampin, and linezolid. The MRSA isolates of other PFGE types were more susceptible to fluoroquinolones, trimethoprim/sulfamethoxazole, as well as gentamicin and were fully susceptible to vancomycin, rifampin, as well as linezolid. The 19 CA-MRSA belonged to 4 PFGE types, C (5 isolates), D (8 isolate), H (4 isolates), and S (2 isolates). Except the 5 isolates of PFGE type C, nearly all other isolates were susceptible to gentamicin, fluoroquinolones, and trimethoprim/sulfamethoxazole with full susceptibility to vancomycin, rifampin, and linezolid. The PFGE patterns were demonstrated in Figure.
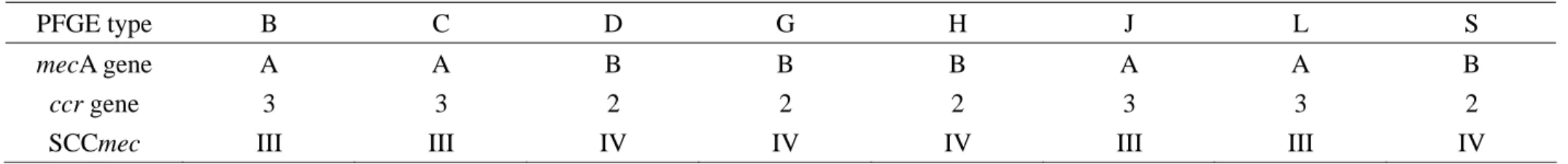
Types of mecA cluster, ccr genes, and SCCmec element:
The types of mecA gene cluster, ccr genes, and SCCmec element of MRSA isolates belonging to PFGE type B, C, D, G, H, J, L, and S were demonstrated in Table 3. Isolates of PFGE type B, C, J, and L carried type III SCCmec element. And isolates of PFGE type D, G, H, and S carried type IV SCCmec element.
討論(含結論與建議)
Our study demonstrated the predominant nosocomial MRSA strain (PFGE type C), which had spread all over Taiwan (10), carried type III SCCmec element and the majority of MRSA isolates isolated in Taiwan carried either type III or type IV SCCmec element. This is a very interesting finding.
Recently, type IV elements were considered to be a marker of CA-MRSA (21). In our present study, 14 out 19 CA-MRSA carried type IV SCCmec element. For the 5 isolates belonging to PFGE type C and classified as CA-MRSA, there were two possibilities. First, the nosocomial strain had been transmitted to community environment. Second, they were just mis-classified and were nosocomial MRSA in fact because that we did not clarify the clinical history in details (patients might have been admitted to another hospital for a period of time before they were admitted to NTUH). In addition, MRSA of PFGE type D accounted for 42.1% of all CA-MRSA. This implied that there was also a predominant MRSA strain, PFGE type C, spread in the community environment in Taiwan.
Among the nosocomial MRSA isolates, there were some isolates (belonging to PFGE type D, G, and H) also carried type IV SCCmec element. This might be due to the transmission and spread of CA-MRSA into hospital environment. However, it was also likely that, in fact, MRSA isolates carried type IV SCCmec element were originated from their ancestor S. aureus isolates which got element and became MRSA in the hospital environment and then transmitted/spread into the community environment. This inference had also been proposed by other researchers (29). However, there was still no solid evidence to verify this hypothesis.
Previous studies had point out that MRSA isolates carring type III SCCmec element were more resistant to antibiotics than MRSA isolates carring other type (especially type IV) SCCmec element (20). Our study also demonstrated the same results. In the present study, MRSA isolates belonging to PFGE type B, C, J, and L, which carried type III SCCmec element, usually were only susceptible to vancomycin, rifampin, and linezolid with little exception. However, MRSA isolates belonging to PFGE type D, G. H, and S, which carried type IV element, usually were susceptible to gentamicin, fluoroquinolone, and
trimethoprim/sulfamethoxazole in addition to rifampin, vancomycin, and linezolid.
In conclusion, our study found that the predominant MRSA strain in Taiwan carried type III SCCmec element and the majority of CA-MRSA carried type IV SCCmec element. There was also a predominant MRSA strain among the CA-MRSA isolates.
Another recent important finding about CA-MRSA was that some CA-MRSA carried Panton-Valentine Leukocidin (PVL) gene (30). The prevalence of PVL gene among CA-MRSA isolates, especially those carried type IV element, in Taiwan was worthy of further study. The origin of so-called CA-MRSA was also another important and interesting issue. Studies on the longitudinal evolution of MRSA in Taiwan might offer help to illuminate this unsolved problem.
參考文獻
1. Jevons MP. ‘Celbenin’-resistant staphylococci. Br Med J 1961;1:124-5. 2. Chang SC, Hsu LY, Luh KT, Hsieh WC. Methicillin-resistant
Staphylococcus aureus infections. J Formos Med Assoc 1988;87:157-63
3. Chang SC, Sun CC, Yang LS, et al. Increasing nosocomial infections of methicillin-resistant Staphylococcus aureus at a teaching hospital in Taiwan.
Int J Antimicrob Agents. 1997;8:109-114.
4. Lee SC, Wang CH, Lee N. Infections caused by methicillin-resistant
Staphylococcus aureus: distinct risk factors and prognosis. Nosocomial Infect Control J (Taiwan). 1996;6:131-138.
5. Ho M, McDonald L, Lauderdale T, et al. Surveillance of antibiotic resistance in Taiwan, 1998. J Microbiol Immunol Infect. 1999;32:239-249.
6. National Nosocomial Infections Surveillance (NNIS) System. (2001). National Nosocomial Infections Surveillance (NNIS) System report: data summary from January 1992-June 2001, issued August 2001. Am J Infect
Control. 2001;29:404-421.
7. Chang SC, Chang HJ, Hsiao ML. Antibiotic usage in public hospitals in Taiwan. J Microbiol Immunol Infect 1998;31:125-32
8. Wang JT, Chang SC, Ko WJ, Chang YY, Chen ML, Pan HJ, Luh KT. A hospital-acquired outbreak of methicillin-resistant Staphylococcus aureus infection initiated by a surgeon carrier. J Hosp Infect 2000;47:104-9 9. Chen ML, Chang SC, Pan HJ, Hsueh PR, Yang LS, Ho SW, Luh KT.
Longitudinal analysis of methicillin-resistant Staphylococcus aureus isolates at a teaching hospital in Taiwan. J Formos Med Assoc 1999;98;426-32. 10. Wang JT, Chen YC, Yang TL, Chang SC. Molecular epidemiology and
antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus in Taiwan. Diagn Microbiol Infect Dis 2002;42:199-203.
11. Song MD, Wachi M, Doi M, Ishino F, Matsuhashi M. Evolution of an inducible penicillin-target protein in methicillin-resistant Staphylococcus
aureus by gene fusion. FEBS lett 1987;221:167-71.
12. Hiramatsu K, Asada K, Suzuki E, Okonogi K, Yokota T. Molecular cloning and nucleotide sequence determination of the regulator region of mecA gene in methicillin-resistant Staphylococcus aureus (MRSA). FEBS lett
1991;298:133-6.
13. Ryffel C, Tesch W, Birch-Machin I, Reynolds PE, Barberis-Makino L, Kayser FH, Berger-Bächi B. Sequence comparison of mecA genes isolated from methicillin-resistant Staphylococcus aureus and Staphylococcus
epidermidis. Gene 1990;94:137-8.
14. Suzuki E, Hiramatsu K, Yokota T. Survey of methicillin-resistant clinical strains of coagulase-negative staphylococci for mecA gene distribution.
Antimicrob Agents Chmother 1992;36:429-34.
15. Suzuki E, Kuwahara-Arai K, Richardson JF, Hiramatsu K. Distribution of
mec regulator genes in methicillin-resistant Staphylococcus clinical strains. Antimicrob Agents Chemother 1993;37:1219-26.
16. Beck WD, Berger-Bächi B, Kayser FH. Additional DNA in and molecular cloning of mec-specific DNA. J Bacteriol 1986;165:373-8.
17. Dubin DT, Matthews PR, Chikramane SG, Stweart PR. Physical mapping of the mec region of an American methicillin-resistant Staphylococcus aureus strain. Antimicrob Agents Chemother 1991;35:1661-5.
18. Ito T, Katayama Y, Hiramatsu K. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant
Staphylococcus aureus N315. Antimicrob Agents Chemother
1999;43:1449-58.
19. Katayama Y, Ito T, Hiramatsu K. A new class of genetic element,
Staphylococcus cassette chromosome mec, encodes methicillin resistance in
Staphylococcus aureus. Antimicrob Agents Chemother 2000;44:1549-55.
20. Ito T, Katayama Y, Asada K, Mori N, Tsutsumimoto K, Tiensasitorn C, Hiramatsu K. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant
Staphylococcus aureus. Antimicrob Agents Chemother 2001;45:1323-36.
21. Ma XX, Ito T, Tiensasitorn C, Jamklang M, Chongtrakool P, Boyle-Vavra S, Daum RS, Hiramatsu K. Novel type of staphylococcal cassette chromosome
mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob Agents Chemother 2002;46:1147-52.
22. National Committee for Clinical Laboratory Standards (NCCLS). Methods
for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, Approved Standard. Villanova, PA: NCCLS, 2000.
23. Bannerman TL, Hancock GA, Tenover FC, Miller JM. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus
aureus. J Clin Microbiol 1995;33:551-5.
24. Jorgensen M, Givney R, Pegler M, Vickery A, Funnell G. Typing
multidrug-resistant Staphylococcus aureus: conflicting epidemiological data produced by genotypic and phenotypic methods clarified by phylogenetic analysis. J Clin Microbiol 1996;34:398-403.
methicillin-resistant Staphylococcus aureus using polymerase chain reaction.
Microbiol Immunol 1992;36:445-53.
26. Matsuhashi M, Song MD, Ishino F, Wachi M, Doi M, Inoue M, Ubukata K, Yamashita N, Konno M. Molecular cloning of the gene of a penicillin binding protein supposed to cause high resistance to beta-lactam antibiotics in Staphylococcus aureus. J Bacteriol 1986;167;975-80.
27. Katayama Y, Ito T, Hiramatsu K. Genetic organization of the chromosome region surrounding mecA in clinical staphylococcal strains: role of
IS431-mediated mecI deletion in expression of resistance in mecA-carring, low-level methicillin-resistant Staphylococcus aureus. Antimicrob Agents
Chemother 2001;45:1955-63.
28. Hiramatsu K, Cui L, Kuroda M, Ito T. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trend Microbiol 2001;9:486-93. 29. de Sousa MA, de Lencastre H. Bridges from hospitals to the laboratory :
genetic portraits of methicillin-resistant Staphylococcus aureus clones.
FEMS Immuno Me Microbio. 2004;40:101-11.
30. Lina G., Piemont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, Vandenesch E, Etienne J. Involvement of Panton-Valentine
leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis 1999;29:1128-1132.
Tabe 1. Drug susceptibility of 31 nosocomial MRSA isolates determined by MIC (μg/mL)
PFGE Oxa Gen Clin Ery Ofl Levo Tet Rif BXT Van Lin
B ≥128 32 ~ 128 ≥128 >128 64 ~ 128 2 ~ 4 128 <0.03 ~ 1 ≥128 1 ~ 2 0.5 C >128 16 ~ >128 ≥128 >128 64 ~ >128 4 ~ 16 64 ~ >128 <0.03 ~ 16 ≥128 0.5 ~ 2 0.125 ~ 1 D >128 0.5 ~ 1.0 ≥128 128 2 ~ 4 0.125 0.125 ~ 16 <0.03 2 0.5 0.25 ~ 1 G 4 ~ 8 0.5 ~ 128 32 ~ ≥128 ≥128 2 ~ 4 0.125 8 ~ 16 <0.03 2 0.125 ~ 0.5 0.25 ~ 1 H 4 ~ 128 1 ~ 128 >128 ≥128 2 ~ 128 0.125 ~ 4 8 ~ 32 <0.03 2 0.25 ~ 0.5 0.5 ~ 1 J ≥128 8 ~ 16 >128 >128 64 ~ 128 2 ~ 4 16 ~ 64 <0.03 >128 0.5 ~ 1 0.5 L >128 64 >128 >128 >128 16 32 ~ 64 16 8 1 0.5
Abbreviation: MIC, minimum inhibitory concentration; Oxa, oxacillin; Gen, gentamicin; Clin, clindamycin; Ery, erythromycin; Ofl, ofloxacin; Levo, levofloxacin; Chl, chloramphenicol; Tet, tetracycline; Rif, rifampin; BXT, trimethoprim/sulfamethoxazole; Van, vancomycin; Lin, linezolid.
Tabe 2. Drug susceptibility of 19 nosocomial MRSA isolates determined by MIC (μg/mL)
PFGE Oxa Gen Clin Ery Ofl Levo Tet Rif BXT Van Lin
C ≥128 16 ~ 32 >128 4 ~ >128 ≥128 4 ~ 16 32 ~ 128 ≦0.03 ≥128 0.5 ~ 1 0.25 ~ 1
D 4 ~ 8 0.25 ~ 1.0 ≥128 8 ~ >128 1 ~ 4 0.06 ~ 0.125 16 ~ 32 <0.03 2 ~ 4 0.5 1
H 8 ~ 16 0.5 >128 64 ~ >128 2 ~ 4 0.125 0.25 ~ 16 <0.03 2 ~ 4 0.5 1
S 4 ~ 16 0.5 >128 ≥128 2 ~ 4 0.125 16 <0.03 2 0.5 1
Abbreviation: MIC, minimum inhibitory concentration; Oxa, oxacillin; Gen, gentamicin; Clin, clindamycin; Ery, erythromycin; Ofl, ofloxacin; Levo, levofloxacin; Chl, chloramphenicol; Tet, tetracycline; Rif, rifampin; BXT, trimethoprim/sulfamethoxazole; Van, vancomycin; Lin, linezolid.
PFGE type B C D G H J L S
mecA gene A A B B B A A B
ccr gene 3 3 2 2 2 3 3 2
SCCmec III III IV IV IV III III IV
計畫成果自評
由上述的研究報告中,可以清楚的發現我們已釐清了台灣地區 MRSA 菌 株,不管是院內或社區性,SCCmec element 的分型;闡明了院內 MRSA 和社區 性 MRSA 所攜帶 SCCmec element 的之差異;發現了社區性 MRSA 菌株也有一個 predominant strain。這些重要的發現,都讓我們對台灣本土的 MRSA 菌株的細菌 特性有了進一步的了解。我們的研究結果,符合原先所設定的個個目標,也提出 了若干進一步的問題、刺激我們進行更深一層的研究。
整體而言,我們的研究結果清楚而重要,不失為台灣本土針對 MRSA 抗藥 性基因的一個好的研究。