國立交通大學
生物科技研究所
碩士論文
克雷白氏肺炎桿菌CG43第三型線毛用於表面抗原呈現與
其表現調控
Application of type 3 fimbriae of Klebsiella pneumoniae
CG43 in epitope display and its expressional regulation
研究生: 楊其駿 (9528505) Student: Chi-Zyun Yang 指導教授: 彭慧玲博士 Adviser: Hwei-Ling Peng, Ph.D
i
中文摘要
為了可以表現克雷白氏肺炎桿菌第三型線毛的重組質體 pmrkABCD 來呈列第二型登革熱病毒的套膜蛋白第三區塊 (DEN2EIII),首先,我們以此包含主要抗原決定位的核酸為模版, 以聚合脢連鎖反應分別增幅可轉譯成約 20 個胺基酸片段長度的核 酸,再將這五段核酸序列分別插入 pmrkABCD 質體中 MrkA A25 或 D27 位置,再將選殖的重組質體包括 pA25-1,pD27-1,pD27-2, pD27-3,pD27-4,pD27-5 分別轉殖到大腸桿菌 JM109。接著,以西 方墨點法分別以 MrkA 和 DEN2EDIII 多株抗體偵測的結果顯示:雖 有不同程度的表現差異,這些重組質體皆可表現帶有 DEN2EDIII 抗 原的重組線毛單體,而此重組的單體蛋白也可組裝成多體的線毛結 構;免疫螢光顯微鏡檢分析也確認這些重組的線毛可表現在細菌表 面。初步純化結果顯示線毛表現的培養條件仍需改進,只有提升線毛 量作為將來免疫小鼠的疫苗來源,才能進一步評估其免疫效果。 大多數的克雷白氏肺炎桿菌臨床分離株帶有可轉錄第三型線毛 的基因組,顯示此線毛在克雷白氏肺炎桿菌附著宿主細胞造成感染過 程中可能扮演重要的角色。至今,細菌如何調控第三型線毛的表現, 仍不清楚。之前的實驗結果顯示,破壞可以調控細菌莢膜產生的調控 基因 rcsB,會增加第一型纖毛的表現,相反的會降低 MrkA 蛋白的表ii
現。我們以帶有 PmrkA-LacZ 報導系統的 CG43S3Z01 的 rcsB 缺損株為
跳躍子接受株,以 X-Gal 培養盤篩選顏色變化,在大約 70000 突變株 中挑出八株會改變顏色的突變珠;其中 6 株會增加 LacZ 活性,2 株 會降低 LacZ 活性。核酸定序分析結果顯示兩株增加 LacZ 活性的插 入點分別是 glycosidase 和 lacI 基因,此二基因與 LacZ 活性調控有關, 與 MrkA 蛋白調控無關。而兩株降低 LacZ 活性的插入點分別是 carbon starvation protein A 和會受到滲透壓影響的 yehZ 基因,此結果確認此 系統的選擇效果,但是跳躍子突變效率仍需提高。
iii
Abstract
We report the use of the type 3 fimbriae, encoded by mrkABCD, from Klebsiella pneumoniae CG43 to display the domain III of envelope protein (E) from type 2 dengue virus, DEN2EDIII. The DNA containing the DEN2EDIII, which has been shown as a major antigenic determinant, was used as a template for PCR amplification to generate 5 DNA
segments (~60 bp) encoding different parts of the DEN2EDIII. The PCR products were cloned into the A25 or D27 site on MrkA reside in the plasmid pmrkABCD. The resulting plasmids including pA25-1, pD27-1, pD27-2, pD27-3, pD27-4 and pD27-5 were then transformed
separatelyinto E. coli JM109. The analysis of these recombinant type 3 fimbriae assessed with western blotting hybridization against anti-MrkA or anti-DEN2EDIII antibody confirmed the expression of the
recombinant proteins, although with different levels. And the
recombinant subunits could assemble into polymeric fimbriae from monomers. The analysis of immunofluorescence microscopy using
anti-MrkA or anti-DEN2EDIII antibody also demonstrated the expression of the recombinant type 3 fimbriae on the surface of the transformed bacteria. However, culture condition and purification method have to be improved to obtain sufficient amount of the recombinant fimbriae. The subsequent use as the vaccine antigen for mouse immunization and the efficacy could then be evaluated.
The fact that most Klebsiella pneumoniae clinical isolates carry type 3 fimbriae gene clusters and hence play an important role as a major adhesin for the bacterial attachment to the host cell to establish an
iv
infection has been speculated. Nevertheless, how the expression of the type 3 fimbriae controlled remains unknown. Herein, a Tn5 mediated mutagenesis was employed to K. pneumoniae CG43Z01rcsB- strain
carrying Pmrk-lacZ. Eight out of approximately 70000 Tn5-insertion mutants
showed alteration of LacZ activity and were isolated. Six mutants
exhibited an increased LacZ activity and two mutants carried a decreased LacZ activity. After the inserted were regions isolated and the sequences determined, it was revealed that the disrupted genes include the gene encoding a putative glycosidase and lacI for the mutants with increasing LacZ activity, and the gene encoding carbon starvation protein and an osmolarity-inducible protein YehZ. The results demonstrated the
feasibility of the reporter-selection and the efficiency of Tn-mutagenesis have to be improved, however.
v 致謝 在這不連續的兩年當中,感謝彭老師的耐心教導。在我有瓶頸 時,不厭其煩地幫助。不論是實驗上的問題,抑或是寫論文做簡報, 老師也都給了我許多的建議。這些幫助,都讓我在往後的研究生涯中 能有很大的助益。也很感謝老師在我出國回來後的繼續教導。 我也感謝丸子學姐一路上耐心的指導我,並且提供我許多建議與 幫助,讓我可以順利地進行我的研究。健誠學長、小新學長與靜柔學 姐也都幫忙我注意實驗上、報告上以及生活上的小細節。哲充也常與 我討論實驗,讓我發現我沒注意到的地方,一些需要體力的事也常常 麻煩他。佩君與家華的開朗,為苦悶的實驗生活帶來很多的歡樂。她 們也常幫忙注意提醒我,讓我不至因為我的不小心的個性而耽誤。志 桓、雅雯、顗峰、小珊、還有朝陽學長也都幫忙我很多。豪君、品瑄 以及崴云也實驗生活提供不少樂趣。 我也感謝我的父母一直以來不斷守護我,忍受我的固執與任性。 還有許許多多的人在我生活上不斷鼓勵我、幫助我、關心我。我很珍 惜這一段日子。
vi
Contents
中文摘要... i Abstract...iii 致謝...iv Contents………vi Abbreviation... viii Introduction...1Materials and methods... 8
Strains, plasmids and growth conditions...………. 8
Recombinant DNA technology...8
Expression of the recombinant fimbriae . ...9
Purification of type 3 fimbriae... 9
Western blotting analysis...10
Immunofluorescence microscopy...11
Preparation of genomic DNA...11
Transposon Mutagenesis...12
Transposon insertion site analysis...12
Results...14
Part I………...14
1. Construction of the DEN2EDIII-MrkA clones...14
2. Assessment of the expression of the recombinant fimbriae………...15
3. Analysis of the expression of the recombinant fimbriae...16
4. Analysis of expression of the recombinant fimbriae with IFM……..16
5. Purification of the recombinant fimbriae………17
Part II……….17
1. Screen for the mutants carrying alteration of MrkA promoter activity………18
2. Analysis of the transposon insertion site………19
Discussion...21
Part I Setup a DEN2EDIII display system using the recombinant type 3 fimbriae……….………...………..…21
Part II Identification of the regulator(s) involved in controlling the expression of type 3 fimbriae………..……… 22
vii
Tables………..………30 Figures………35
viii
Abbreviation
cfu Colony-forming unit
DENIIDIII Dengue virus serotype II domain III
ELISA The Enzyme-Linked ImmunoSorbent Assay FITC Fluorescein isothiocyanate
g The acceleration due to gravity at the earth’s surface IgG Immunoglobulin G kb Kilobase(s) kDa Kilodalton(s) g Microgram ml Milliliter l Microliter mm Millimeter mM Millimolar M Micromolar
MR/K Mannose resistance Klebsiella-like hemagglutinins nm Nanometer
OD Optical Density
PAGE Polyacrylamide gel electrophoresis PBS Phosphate-Buffer saline
PCR Polymerase chain reaction SDS Sodium dodecyl sulfate
1
Introduction
Klebsiellap neumoniae, an Enterobacteriaceae gram negative bacterium, is recognized as an important opportunistic pathogen that attacks immunocompromised patients. It is a common cause of urinary tract infections, respiratory tract infections and septicaemia especially in immunocompromised individuals who are hospitalized and suffer from severe underlying diseases, such as chronic pulmonary obstruction or diabetes mellitus (Chen, Hsueh et al. 2000). In recent years, the cases of diabetes mellitus patients that suffer from liver abscess (LA) caused by highly virulent K. pneumoniae strains have increasingly occurred.
Several virulence factors of K. pneuniornae have been identified, including acidic polysaccharide capsule (CPS), lipopolysaccharides, iron acquisition systems and several distinct types of adherence factors (Nassif and Sansonetti 1986). The virulence traits for KP-PLA (pyogenic liver abscess)have recently been reported, which include K1, K2, magA, rmpA (regulator of mucoid phenotype), uge (UDP-glucose4-epimerase), wabG, and iron acquisition systems encoded by iuc and iro, kfu (an iron uptake system), and TonB (Izquierdo, Coderch et al. 2003; Regue, Hita et al. 2004; Hsieh, Lin et al. 2008). The capsular serotype K1 or K2 has been shown to be more prevalent in strains causing liver abscess than the ones causing bacteremia(Yeh, Kurup et al. 2007). The magA gene located in the K1 cps gene cluster has been reported in 98.1% to 83.3% of K. pneumoniae strains isolated from patients with LA and was significantly more prevalent than the bacteremic strains. Mutations resulting the loss of K1 CPS became avirulent in mouse model (Chuang, Fang et al. 2006).
2
The prevalence of rmpA in LA strains was also higher than that in bacteremic strains. The iron uptake system encoding genes have been shown to be highly up-regulated in response to sera. Nevertheless, mutation in all of the 3 loci (irp2, iuc, and iroA) is necessary to decrease the virulence (Huang, Liao et al. 2009). The tonB mutant has been shown to be a potential vaccine candidate because it can induce a significant protective immune response (Huang, Liao et al. 2009).
Adherence factors which enable bacterial binding to specific host cells play important roles in establishing an infection. Fimbriae, one of the adherence factors, are long, thread-like appendages on bacterial surface. They are found in as many as 500 copies per cell (Klemm and Schembri 2000). Each fimbrial fiber is a polymer composed of hundreds of structural subunits called pilin. In the infection process, the adhesin which is located on the tip of fimbriae determines the specific binding to the host cell.
Most clinical K. pneumoniae isolates express two types of fimbrial adhesins, type 1 and type 3 fimbriae (Gerlach, Clegg et al. 1989). Type 1 fimbriae found in a majority of enterobacterial species are about 7 nm wide and 1 m long. Activity of type 1 fimbriae was determined by their ability to mediate mannose-sensitive agglutination of guinea pig or fowl erythrocytes, and hence referred as mannose-sensitive hemagglutinins (MSHA). They have been shown to be required for bacterial attachment to mannose units of the glycoprotein receptor on the surface of urinary epithelium cells(Buchanan, Falkow et al. 1985). A number of studies
3
have shown that type 1 fimbriae play a significant role in the ability of Escherichia coli infection of the urinary tract(Connell, Agace et al. 1996). Recently, type 1 fimbriae were also established as an important virulence factor in K. pneumoniae urinary tract infection (Struve, Bojer et al. 2009).
Type 1 fimbriae, expressed by fimACDFGHIK gene cluster, are composed primarily of the structural subunit FimA, with minor amounts of three ancillary subunits, FimF, FimG, and the mannose-specific
adhesin FimH. FimH adhesin is an allosteric protein that mediates the catch bond mechanism of adhesion where the binding is increased under increased shear stress. The fimC and fimD genes respectively encode a fimbrial chaperone and usher protein.The function of fimI gene product is unknown, but this product has been found to be essential for type 1
fimbria biosynthesis in E. coli (Valenski, Harris et al. 2003). The fimK gene located directly downstream of fimH is only present in K.
pneumoniae but not in E. coli. The fimK gene product has previously been shown to be involved in type 1 fimbria expression (Rosen, Pinkner et al. 2008). Regulation of the type 1 fimbria expression in E. coli is very complex, and several regulatory factors that act by altering the expression of fimB and fimE have been described. The recombinases FimB and FimE regulate the phase switch of type 1 fimbriae. FimE turns the switch from “ON” position to “OFF” position, while FimB can turn the switch in either direction(Klemm 1986).
Type 3 fimbriae are 2 to 4 nm wide and 0.5 to 2 m long fimbriae (Hornick, Thommandru et al. 1995). They are characterized by their
4
ability to agglutinate tannic acid treated, but not native, erythrocytes in a mannose resistant manner, and hence they are referred as mannose resistant Klebsiella-like (MR/K) hemagglutinin (Duguid 1959; Gerlach, Clegg et al. 1989). Besides Klebsiella species, type 3 fimbriae are common in the isolates of Enterobacter, Serratia, Proteus and
Providencia (Old and Scott 1981; Old and Adegbola 1982; Korhonen, Tarkka et al. 1983; Buchanan, Falkow et al. 1985; Old and Adegbola 1985). Historically, type 3 fimbriae have not been associated with E. coil; however, two recent independent studies have reported the expression of type 3 fimbriae in E. coil strains (Ong, Ulett et al. 2008). In vitro studies have revealed that type 3 fimbriae mediate adhesion to different
structures in human kidney and lung tissue, epithelial cells from human urine sediments as well as endothelial and bladder epithelial cell lines (Struve, Bojer et al. 2009). Furthermore, type 3 fimbriae of K.
pneunioniae have been shown to influence the biofilm formation (Struve, Bojer et al. 2009). Their involvement of biofilm formation and binding to different host structures suggested an important role in determining
bacterial virulence.
Type 3 fimbriae are encoded by the mrk gene cluster, which includes the mrkA gene encoding the major fimbrial subunit and mrkD encoding the fimbrial adhesin responsible for MR/K hemagglutination. It has been reported that the N terminal domain of MrkD adhesin is responsible for receptor binding (Tarkkanen, Allen et al. 1990). MrkD adhesin has also been shown to mediate adhesion to collagen; however, the exact identity of the MrkD receptor remains elusive (Hornick, Thommandru et al. 1995).
5
MrkC is an outer membrane usher protein, which anchors the fimbriae to the bacterial cell surface. MrkB, a periplasmic chaperone, acts to stabilize fimbrial subunits and carries them to the outer membrane usher
protein(Choudhury, Thompson et al. 1999). Downstream of mrkD is mrkF of which the encoding product has been reported to affect stability of the intact fimbrial appendages on bacterial surface (Huang, Liao et al. 2009).
Assembly of type 3 fimbriae is via the chaperone-usher pathway, in which the chaperone-mediated extraction of subunits from the inner cytoplasmic membrane (IM) is coupled with their folding into an assembly-competent state(Sauer, Barnhart et al. 2000). The
immunoglobulin-like chaperones protect the nascent-folded subunits from premature oligomerization in the periplasmic space by directly capping. These preassembly complex protected by the chaperone was then
delivered to the outer membrane (OM) assembly site that is comprised of the usher.
Analysis of the gene loci upstream of the mrkABCDF revealed genes homologous to pecM and pecS, which encode regulators for the
expression of virulence genes in Erwinia chrysanthemi (Reverchon, Nasser et al. 1994). As shown in Fig. 1, the two divergently transcribed genes were named phgM and phgS respectively (黃盈蓉 2006). The genes downstream to the mrkABCEF are genes KP4551, KP4552, and KP4554 encoding c-di-GMP binding domain protein (PilZ domain protein), transcription factor with an HTH domain, and c-di-GMP
6
phosphodiesterase (EAL) domain protein (Fig. 2), respectively. Whether the regulators are involved in the control of the expression of type 3 fimbriae remains unknown.
Fimbriae which expressed on bacterial surface have been evaluated as a target protein display system for medicine selection or vaccine development. Fimbriae are particularly attractive candidates for epitope expression and several of the properties also hold promise for the
feasibility of vaccine development: first, fimbriae are present in extremely high numbers at the cell surface; second, they are as strong immunogens, and thirdly, they can be easily purified(Connell, Agace et al. 1996). Until now, hepatitis B virus and foot-and-mouth disease virus epitopes have been successfully expressed on type 1 fimbriae and P pili of E. coli (Hedegaard and Klemm 1989; Bakker, van Zijderveld et al. 1990).
Dengue virus (DV) is an arthropod-borne human pathogen that causes a serious public health threat in tropical and subtropical regions of the world. DV has four serotypes (DEN-1 to DEN-4) that cause diseases ranging from mild dengue fever to severe symptoms such as dengue hemorrhagic fever and dengue shock syndrome (Chambers, Hahn et al. 1990). The virion of DEN contains three structural proteins: a 12 kDa nucleocapsid or core protein (C), 8 kDa membrane protein (M), and 53 kDa envelope protein (E), as well as seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, NS5). The X-ray structures reveal that the E protein is composed of three domains, domain I, II, and III. The
7
domain III, an immunoglobin(Ig)-like domain, is considered as a location that bind with host cell receptor(Konishi, Kosugi et al. 2006). The recent report employing the domain III-specific monoclonal antibody has
demonstrated a neutralization activity to block the viral infection(Modis, Ogata et al. 2005).
In this thesis, two studies are carried out. The first one is to “Set up a DEN2-EIII display system using type 3 fimbriae”. We plan to generate recombinant type 3 fimbriae by introducing DEN2-EIII amino acid sequences to MrkA pilin and then purify the recombinant fimbriae to immunize mouse to evaluate the possibility as vaccine antigen. The
second aim is to “Identify possible regulator(s) involved in controlling the expression of type 3 fimbriae”. We use Tn5 transposon, which can
randomly insert into to the chromosome of the K. pneumoniae harboring the PmrkA-LacZ reporter plasmid (Fig. 3). The assay takes advantage of the
fact that-galactosidase activity could be monitored by the changes of white/blue color on the selective medium containing X-gal.
8
Materials and Methods
Strains, plasmids and growth conditions
The bacteria strains, plasmids and primers used in this study are listed in Tables 1, 2, and 3. Bacteria were incubated at 37°Cin Luria Broth (LB) medium or the medium supplemented with antibiotic ampicillin (100 g/ml), kanamycin (25 g/ml) or chloramphinicol (35
g/ml). The bacterial growth was determined by measuring the optical density at 600 nm (OD600).
Construction of the recombinant plasmids
The recombinant plasmids pD27-1, pD27-2, pD27-3, pD27-4, pD27-5, and pA25-2 clones were constructed by PCR cloning using pGEM-T Easy-D2E as the template, and the 5 amplified fragments cloned respectively into yT&A cloning vector. The PmeI was then used to cleave the clone and the PmeI fragment, containing the target DNA was respectively subcloned into A25 and D27 site of pMrkABCD (徐幸 瑜 2007).
9
Bacteria were incubated at 37°C in Luria Broth (LB) overnight and collected by centrifugation for 1 min (8,000 rpm). The pellet was
resuspended in 150 l protein lysis buffer (50 mM Tris-HC1 pH 8.0, 1 mM EDTA, 100 mM NaC1), added with 15 l of 4% SDS loading dye (100 mM Tris-HCl pH 8.0, 4% SDS, 0.2% bromophenol blue, 200 mM
-mercaptoethanol, 20% glycerol), and then heated at 95°C for 30 min. The cell extracts were resolved using 13.5% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Protein expression for fimbriae polymer pattern
Bacteria were incubated at 37°C in Luria Broth (LB) overnight and collected by centrifugation for 1 min (8,000 rpm). The pellet was
resuspended in 150 l protein lysis buffer (50 mM Tris-HC1 pH 8.0, 1 mM EDTA, 100 mM NaC1), added with 15 l of 4% SDS loading dye (100 mM Tris-HCl pH 8.0, 4% SDS, 0.2% bromophenol blue, 200 mM
-mercaptoethanol, 20% glycerol). The cell extracts were resolved using 13.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis
(SDS-PAGE).
Purification of type 3 fimbriae
The fimbriae purification process was carried out with a minor modification of the described methods (Feutrier, Kay et al. 1986;
10
Girardeau, Der Vartanian et al. 1988; Sakellaris, Balding et al. 1996; Zalewska, Piatek et al. 2003). The recombinant E. coli JM109 [D27], E. coli JM109 [D27-1], E. coli JM109 [D27-2], E. coli JM109 [D27-3], E. coli JM109 [D27-4], E. coli JM109 [D27-5], E. coli JM109 [A29-2], and JM109[pmrkABCD] were cultured at 37°Cfor 24 h in 24 ml LB broth, and the bacteria were collected by centrifugation for 3 min (8,000 rpm, 4°C) and were suspended in 1 ml phosphate-buffered saline (PBS). The bacterial suspension was heated at 65°C for 2 h and homogenized in blender for 20 min at ambient temperature. Finally, 30% saturated ammonium sulfate at 4°C was added and the mixture stored at 4°C
overnight. The fimbriae containing pellet was collected by centrifugation (20,000 g, 90 min) and suspended in 40 l PBS.
Western blotting analysis
The fimbrial proteins were resolved by 13.5% (for fimbriae
monomer pattern) or 8% (for fimbriae polymer pattern) SDS-PAGE, and electrophoretically transferred to polyvinylidene difluoride membranes (ImmobilonTM-P, Millipore). Subsequently, the membranes were soaked in 5% skim milk in 1 x phosphate-buffered saline (PBS) at 4°C overnight. After washing 3 times with 1 x PBS, the membrane was incubated with the antiserum of a 10000-fold diluted anti-MrkA or 5000-fold diluted anti-DEN2EDIII at room temperature for 2 h. Followed by incubation with a 5000-fold diluted alkaline phosphatase-conjugated anti-rabbit immunoglobulin at room temperature for 2 h, three washes were applied
11
and the bound antibodies were detected by using the chromogenic
reagents BCIP (5-bromo-4-chloro-3-indolyl phosphate) and NBT (Nitro blue tetrazolium).
Immunofluorescence microscopy
Bacteria cultured in LB at 37°C for 16 h were collected, washed twice by phosphate buffer saline (PBS, 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.3), and were dissolved in 1 ml of sterile water. Ten microliters of the bacterialsuspension were loaded on glass slide and the bacteria thermally-fixed. Subsequently, an appropriate concentration of diluted antibody (1/300 of anti-MrkA or 1/5 dilution of anti-DEN2DIII) was added and incubated at room temperature 1 h, and then washed twice by PBS. An appropriate dilution of secondary
antibody (1/100 of the FITC conjugated anti-rabbit IgG) was then added and the slide incubated at room temperature for 1 h in dark. After washed twice with PBS, a drop of 10% glycerol phosphate buffer was added on the slide. Finally, the slide was covered with coverslip, fixed with nail polish, and placed under the fluorescent microscope for observation.
Preparation of genomic DNA
The bacteria cultured at 37°C in LB overnight were collected by centrifugation for 3 min (8,000 rpm). The pellet was resuspended in 800
12
EDTA, 40 mM Tris-HCl pH 8.0, 0.2% Triton X-100) and heated at 37℃ for 30 min. Proteinase K solution 100 g/ml was then added and heated at 50°C overnight. The solution was put on ice for 10 min and 250l saturated NaCl solution was added, and the carefully mixed by shaking for 10 min. After centrifugation for 20 min (13000 rpm), 500 l of the supernatant was added with 1000 l 99% alcohol in an eppendorf tube. The precipitate collected by centrifugation for 10 min (13000 rpm). After gently rinsed with 75% alcohol, the pellet was dried and resuspended in sterile water.
Transposon Mutagenesis
Transposon mutagenesis with a mini-Tn5(Km) was performed by biparental mating (de Lorenzo, Herrero et al. 1990). The recipient (K. pneumoniae CG43Z01 rcsB- [P-mrkA-LacZ]), donor (E. coli S17-1 λ pir
[pUTmini-Tn5Km2]), were grown overnight in LB with the appropriate antibiotics. After incubation of the recipient and donor at 37°C for 30 min, 0.5 ml of the recipient was mixed with 1 ml of the donor. Cells were
collected by centrifugation, suspended in 100 l of saline solution, and spotted on an NC membrane on LB plate. After 16 h incubation at 37°C, cells were scraped off the NC membrane and resuspended in 5 ml of LB, and serial dilutions were plated on selective minimal medium, which was M9 with kanamycin and chloramphenicol.
13
In order to analyze the Tn-insertion sites, chromosomal DNA of the mutants containing the mini-Tn5 Km2 were isolated and then subjected to restrict enzyme PstI digestion. The restricted fragments were then ligated to the PstI site of pUC19 and the ligation mixture used to transform to E. coli JM109. The transformants were selected using the LB plates
supplemented with both kanamycin and ampicillin. Finally, the
14
Results
Part I
Setup a DEN2EDIII display system using type 3 fimbriae
1. Construction of the DEN2EDIII-MrkA clones
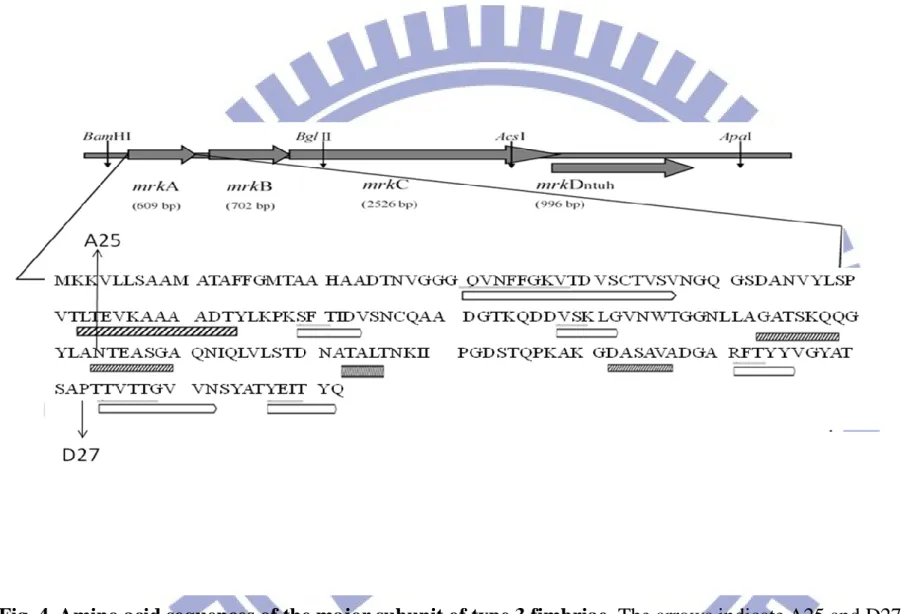
It has been shown that the insertion of a small peptide in the residue A25 or D27 of MrkA (Fig.4) had no apparent effect on the expression of the recombinant fimbriae (徐幸瑜 2007). Whether the insertion of
DEN2EDIII affects the expression of the fimbriae was investigated. The recent report employing the monoclonal antibody specific to DEN2E domain III has demonstrated a neutralization activity(Abd-Jamil, Cheah et al. 2008). In order to use type 3 fimbriae as an expression
system for the vaccine development, selection of an epitope-containing peptide is important. It is hence the DEN2EDIII was selected as a foreign sequence to be displayed by the type 3 fimbriae expression system which has previously been set up in our laboratory (徐幸瑜 2007).
It has been reported that the major subunit of type 1 fimbriae can accommodate the size of the protein fragments ranging between 10-30 amino acids(Klemm and Schembri 2000). As shown in Fig. 5A, the ~ 100 aa DEN2EDIII divided into 5 fragments of about 20 amino acids (Fig. 5B) was cloned by PCR. The PCR products were cloned into yt&A, and then subcloned using the restriction enzyme PmeI to the plasmid pA25 and pD27, respectively. Six recombinant plasmids were obtained and named
15
pA25-2, pD27-1, pD27-2, pD27-3, pD27-4 and pD27-5.
As shown in Table 4, sequence analysis of these recombinant clones revealed additional amino sequences, which derived from the in-frame construct on the designed primer sequences added to the DEN2EDIII fragments. The resulting clones A25-2, D27-1and D27-2 contained 29 aa; 28 aa for D27-3; 30 aa for D27-4 and D27-5. Notably, the D27-3
contained some alterations of the DEN2EDIII fragment 3 sequence.
2. Assessments of the expression of the recombinant DEN2EDIII-MrkA fimbriae
In order to investigate whether the insertion of DEN2EDIII fragments affect the expression of the fimbriae, total lysates from the recombinant E. coli JM109 containing A25-2, D27-1, D27-2, D27-3, D27-4 or D27-5 were collected and the proteins analyzed by SDS-PAGE and western blotting analysis. Since the recombinant MrkA of the
displayed vector pD27 carries 5 extra aa (徐幸瑜 2007), higher
molecular weight than ~ 23 kDa of the MrkA was expected (lanes 2 and 3 of Fig. 6A). Except the recombinant protein of D27-4, the molecular weight of the other recombinant MrkA also appeared to be in larger size than the native MrkA. As shown in Fig. 6A, extra band with
approximately 2 kDa larger, which is probably corresponding to the signal peptide-containing recombinant MrkA, were observed.
Expression of the recombinant MrkA was subsequently demonstrated by western blotting analysis with anti-MrkA antibody. As shown in Fig.
16
6B, the arrowhead-labeled bands in Fig. 6A could be hybridized with the anti-MrkA antibody indicating expression of the recombinant MrkA. The insertion of the DEN2EDIII fragments had no apparent effect on the
expression.Whether the DEN2EDIII fragments were properly expressed was also investigated. As shown in Fig. 6C, western blotting analysis using anti-DEN2EDIII antibody showed only the recombinant clones carrying MrkA-DEN2EDIII could be detected using anti- DEN2EDIII antibody.The detection for D27-3 and D27-5 (lanes 7 and 9, respectively) exhibited higher intensity than those of the others implying a high level expression of the DEN2EDIII fragments.
3. Analysis of the expression of the recombinant fimbriae
Polymer pattern analysis for the recombinant clones was carried out to study if the recombinant DEN2EDIII-MrkA monomer could be
assembled properly onto the bacterial surface. As shown in Fig. 7A, approximately same amount of the total proteins were resolved on the SDS-polyacryamide gel without heat treatment.The western blotting analysis with anti-MrkA antibody revealed different levels of polymer formation, and the polymeric fimbriae of D27-4 appeared to be better formed than the others (Fig. 7B).
4. Analysis of expression of the recombinant fimbriae with IFM
IFM analysisprobed with anti-MrkA or anti-DEN2EDIII antibody was also applied to confirm the expression of the recombinant fimbriae
17
on the bacterial surface. As shown in Fig.8A, all the recombinant fimbriae carrying with the recombinant MrkA could be observed indicating
expression on the cell surface. The analysis using anti-DEN2EDIII as shown in Fig. 8B, expression of the recombinant fimbriae was confirmed. Although the inserted DEN2EDIII fragments contain only about 25
amino acids, the florescent signals were apparent implying proper expression of the DEN2EDIII peptides.
5. Purification of the recombinant fimbriae
In order to use the fimbriae to induce effective animal immune
reaction, a quick and simple purification method is essential. To avoid the process of ultracentrifugation, we’ve tried to modify the purification
procedures according to the methods (Feutrier, Kay et al. 1986; Girardeau, Der Vartanian et al. 1988; Sakellaris, Balding et al. 1996; Zalewska,
Piatek et al. 2003). Analysis of the purified fimbriae as shown in Fig. 9A revealed that most of the recombinant fimbriae were in short oligomeric form. The western blotting analysis indicated that the purified
recombinant fimbriae from the D27-4 strain had similar polymeric pattern, although with lower amount, as those from pmrkABCE or pD27-4 (Fig. 9B). The barely detectable bandings of the other recombinant fimbriae indicated a poor purification result.
Part II
Identification of the regulator(s) involved in controlling the expression of type 3 fimbriae
18
RcsB is a two-component regulator of capsule synthesis (Stout and Gottesman 1990). It has been recently reported in E. coli that the rcsB deletion decreased the expression of type 1 fimbriae suggesting a positive regulatory role for the expression of type 1 fimbriae (Schwan, Shibata et al. 2007). In contrast, our laboratory has shown that the deletion of rcsB in K. pneumoniae CG43 increased the expression of type 1 fimbriae. However, the expression of MrkA, the major subunit of type 3 fimbriae was reduced (data not shown). Since the regulatory system for the expression of type 3 fimbriae remains unknown, we intend to apply transponson mutagenesis onto the rcsB deletion mutant for a possible suppression of the phenotype. To facilitate the screening process, the promoter reporter construct was employed. It is hence total proteins isolated from K. pneumoniae CG43S3Z01 and CG43S3Z01rcsB-, and CG43S3Z01rcsB-[pLacZ15] and CG43S3Z01rcsB-[pPmrkA-lacZ] were
comparatively analyzed to confirm the rcsB deleting effect. As shown in Fig. 10A, approximately similar amount of the proteins were isolated. The subsequent western blotting analysis using anti-MrkA antibody showed that the rcsB deletion indeed reduced the expression of MrkA of CG43S3Z01rcsB- or CG43S3Z01rcsB-[pPmrkA-lacZ].
1. Screen for the mutants carrying alteration of MrkA promoter activity
As shown in Fig.3, Tn5 mobilized from E. coli to K. pneumoniae CG43S3Z01 rcsB-[pPmrkA-lacZ] and the transconjugants were selected on
19
X-gal containing M9-Km plates. Approximately 70,000 mutants were picked onto LB-X-gal plate to further clarify the changes of the colony color. Finally, eight mutants with apparent color changes, two white and six deep blue, were isolated and named M1~M8 individually.
2. Analysis of the transposon insertion site
In order to determine the inserted sequences, chromosomes of the eight mutant were isolated and completely digested by restriction enzyme PstI. As shown in Fig. 3, there is a PstI site outside the transposon kanamycin-resistant gene and hence the nucleotides of K. pneumoniae next to the kanamycin-resistant gene could be included with PstI digestion. The M4 strain grew poorly on the kanamycin/ chloramphinicol- containing medium and hence was not included for the following study. The PstI-digested fragments were cloned into vector pUC19, and the transformants selected on Knanmycin containing plates. As shown in Fig11B, except M3, six recombinant clones namely pM1, pM2, pM5, pM6, pM7, and pM8 were obtained and further confirmed by PstI digestion. After subjected to more insertion enzymes digestion, the restriction pattern of pM1 and pM6 appeared to be identical (Fig. 11C). Finally, the clones were sent to commercial service for sequence determination using the primers M13R and M13F.
The Blast tool (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi) was then applied to analyze the determined sequences and the inserted genes by the transposon include a putative glycosidase encoding gene (Fig. 12A), lacI (Fig. 12B), gene encodes RelE/ParE family or carbon
20
starvation protein A (Fig. 12C), and yehZ gene (Fig. 12D). YehZ is homologous to OsmF, an osmotically inducible protein (Checroun and Gutierrez 2004).
21
Discussion
Part I
Setup a DEN2EDIII display system using the recombinant type 3 fimbriae
Analysis of monomeric form of the recombinant MrkA indicated that the insertion of about 30 amino acids of the DEN2EDIII fragments had no apparent influence on the expression of MrkA. However, unexpected protein size of D27-3 (Fig. 6A, lane 7) and D27-4 (Fig. 6A, lane 8) were found. Analysis of pD27-3 revealed that two DEN2EDIII fragment 3 were inserted into MrkA and hence size of the recombinant MrkA was the largest. This also suggested that the insertion sequence could be up to 47 aa with no apparent effect on the expression of the recombinant MrkA. On the other hand, the insertion of part 4 of DEN2EDIII may alter the structure of MrkA and hence resulted in a smaller protein in D27-4 (lane 8 in Fig. 6A).
The upper band of the two major bands in Fig. 6A is speculated as the one still carrying the signal peptide as a cytosolic form. As shown in the lane 2 and lane 8 in Fig 6A, one major band was observed indicating that most of the MrkA had been transported out to the periplasmic space. The differential transported level is probably due to different exposure of the signal peptide on the recombinant MrkA to be recognized by the signal peptidase.
22
antibody, the polymer pattern analysis revealed only the recombinant fimbriae D27-4 could be comparable with that of the wild type carrying pmrkABCD. This is probably due to the smallest size of the monomeric form of D27-4 could be better assembly into polymeric form. As shown in Fig.8, the better detection of D27-3 and D27-5 by IFM analysis indicated the two recombinant epitopes are properly exposed to be detected by anti-DEN2EDIII antibody.
The previous study has shown that the recombinant type 3 fimbriae inserted PERV (Porcine endogenous retrovirus) envelope amino acid sequence can inducePERV envelope antibody in mouse (陳欣瑜 2008). We’ve expected that the purified recombinant fimbriae can induce immune reaction in animals as well. However, the purified fimbriae, even from D27-4 (lane 8 in Fig.9A) with the largest amount, were not enough for the following immunization study. Since the purified fimbriae appeared to be good, alternative way such as improving the cultured condition for stable fimbrial expression might be helpful to obtain enough amounts of the fimbriae for immunization.
Part II
Identification of the regulator(s) involved in controlling the expression of type 3 fimbriae
Fimbriae are an important pathogenic factor for bacteria to attach to the host cell. Regulation to control the expression of the fimbriae is conceivably important. Herein, the random mutagenesis employed had identified several genes which may have a role for the expression of type
23
3 fimbriae. The analysis of transposon insertion site of M2 mutant revealed a putative glycosidase. Glycosidase (also called Glycoside hydrolase), catalyzes the hydrolysis of the glycosidic linkage to generate two smaller sugars, has been found in essentially all domains of life. In bacteria and prokaryotes, glycosidase are found both as intracellular and extracellular enzymes that are largely involved in nutrient acquisition (Dwek 1996). The enzyme beta-galactosidase (LacZ) is one of the important occurrences of glycoside hydrolases in bacteria. The white colony of M2 mutant could be resulted from the disruption of the glycosidase gene and hence affected the bacterial digestion of X-gal. The insertion site of M5 was mapped to lacI gene. This could be easily explained by the inability of LacI to repress the LacZ activity and hence a deep blue colony was observed.
Since the available sequences of the M7 clone contained only K. pneumonia genes, the transposon insertion site was speculated. As shown in Fig. 12C, the disrupted genes could be the gene encoding plasmid stabilization system protein of RelE/ParE family or carbon starvation protein A, CstA. RelE/ParE is a family of TA systems, which typically consist of pairs of genes: one for a stable toxin that can cause cell death by disrupting an essential cellular process and the other for a labile antitoxin that can bind the toxin and block activity of the toxin (Anantharaman and Aravind 2003; Engelberg-Kulka, Amitai et al. 2006). The deletion of TA systems has been shown to decrease biofilm formation initially (8 h) on three different surfaces and then increased biofilm formation (24 h) by decreasing biofilm dispersal (Kim, Wang et al. 2009).
24
CstA is involved in peptide transport that would assist the cell in escaping carbon starvation (Schultz and Matin 1991). If disruption of the relE/parE gene or cstA could influence the MrkA expression remains to be investigated.
The disrupted gene of M8 is yehZ gene which is located within yehZYXW operon and coding for a putative glycine/betaine/choline transport protein (ABC superfamily). The yehZYXW encoded ABC transporter as an additional element of the global stress response controlled by sigma(s) (Checroun and Gutierrez 2004). The analysis of a yehZ-lacZ transcriptional fusion demonstrated that yehZ is inducible not only by osmolarity, but also upon entry into stationary phase. CstA and YehZ both are related with membrane transporters. Whether they have influences on MrkA expression awaits to be clarified.
While sequencing the other mutant clones, transposase sequence has been identified indicating some unexpected recombination events had occurred. How to improve the transposon delivery system has to be taken into consideration.
25
Reference
黃盈蓉. (2006). Characterization of type 3 fimbriae in Klebsiella pneumoniae. 國立交通大學博士學位論文
徐幸瑜.(2007). MrkA, the major pilin of Klebsiella pneumoniae type3 fimbriae — identification of critical regions for the fimbriae assembly and dispensable regions for display system development. 國立清華大學碩士學位論文
陳欣瑜.(2008). MrkA, the major pilin of Klebsiella pneumoniae type 3 fimbriae ─ Identification of the important residues involved in pilus assembly and construction of a vaccine display system. 國立清華大學碩士學位論文
Abd-Jamil, J., C. Y. Cheah, et al. (2008). "Dengue virus type 2 envelope protein displayed as recombinant phage attachment protein reveals potential cell binding sites." Protein Eng Des Sel 21(10): 605-611.
Anantharaman, V. and L. Aravind (2003). "New connections in the prokaryotic toxin-antitoxin network: relationship with the eukaryotic nonsense-mediated RNA decay system." Genome Biol 4(12): R81.
Bakker, D., F. G. van Zijderveld, et al. (1990). "K88 fimbriae as carriers of heterologous antigenic determinants." Microb Pathog 8(5): 343-352.
Buchanan, K., S. Falkow, et al. (1985). "Frequency among
Enterobacteriaceae of the DNA sequences encoding type 1 pili." J Bacteriol 162(2): 799-803.
Chambers, T. J., C. S. Hahn, et al. (1990). "Flavivirus genome organization, expression, and replication." Annu Rev Microbiol 44: 649-688.
Checroun, C. and C. Gutierrez (2004). "Sigma(s)-dependent regulation of yehZYXW, which encodes a putative
26
osmoprotectant ABC transporter of Escherichia coli." FEMS Microbiol Lett 236(2): 221-226.
Chen, K. Y., P. R. Hsueh, et al. (2000). "A 10-year experience with bacteriology of acute thoracic empyema: emphasis on
Klebsiella pneumoniae in patients with diabetes mellitus." Chest 117(6): 1685-1689.
Choudhury, D., A. Thompson, et al. (1999). "X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli." Science 285(5430): 1061-1066.
Chuang, Y. P., C. T. Fang, et al. (2006). "Genetic determinants of capsular serotype K1 of Klebsiella pneumoniae causing
primary pyogenic liver abscess." J Infect Dis 193(5): 645-654. Connell, I., W. Agace, et al. (1996). "Type 1 fimbrial expression
enhances Escherichia coli virulence for the urinary tract." Proc Natl Acad Sci U S A 93(18): 9827-9832.
de Lorenzo, V., M. Herrero, et al. (1990). "Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria." J Bacteriol 172(11): 6568-6572.
Duguid, J. P. (1959). "Fimbriae and adhesive properties in Klebsiella strains." J Gen Microbiol 21: 271-286.
Dwek, R. A. (1996). "Glycobiology: Toward Understanding the Function of Sugars." Chem Rev 96(2): 683-720.
Engelberg-Kulka, H., S. Amitai, et al. (2006). "Bacterial programmed cell death and multicellular behavior in bacteria." PLoS Genet
2(10): e135.
Feutrier, J., W. W. Kay, et al. (1986). "Purification and
characterization of fimbriae from Salmonella enteritidis." J Bacteriol 168(1): 221-227.
Gerlach, G. F., S. Clegg, et al. (1989). "Identification and
characterization of the genes encoding the type 3 and type 1 fimbrial adhesins of Klebsiella pneumoniae." J Bacteriol
171(3): 1262-1270.
Girardeau, J. P., M. Der Vartanian, et al. (1988). "CS31A, a new K88-related fimbrial antigen on bovine enterotoxigenic and septicemic Escherichia coli strains." Infect Immun 56(8): 2180-2188.
27
Hedegaard, L. and P. Klemm (1989). "Type 1 fimbriae of Escherichia coli as carriers of heterologous antigenic sequences." Gene 85(1): 115-124.
Hornick, D. B., J. Thommandru, et al. (1995). "Adherence properties of an mrkD-negative mutant of Klebsiella pneumoniae." Infect Immun 63(5): 2026-2032.
Hsieh, P. F., T. L. Lin, et al. (2008). "Serum-induced iron-acquisition systems and TonB contribute to virulence in Klebsiella
pneumoniae causing primary pyogenic liver abscess." J Infect Dis 197(12): 1717-1727.
Huang, Y. J., H. W. Liao, et al. (2009). "MrkF is a component of type 3 fimbriae in Klebsiella pneumoniae." Res Microbiol 160(1): 71-79.
Izquierdo, L., N. Coderch, et al. (2003). "The Klebsiella pneumoniae wabG gene: role in biosynthesis of the core lipopolysaccharide and virulence." J Bacteriol 185(24): 7213-7221.
Kim, Y., X. Wang, et al. (2009). "Toxin-antitoxin systems in Escherichia coli influence biofilm formation through YjgK (TabA) and fimbriae." J Bacteriol 191(4): 1258-1267. Klemm, P. (1986). "Two regulatory fim genes, fimB and fimE,
control the phase variation of type 1 fimbriae in Escherichia coli." EMBO J 5(6): 1389-1393.
Klemm, P. and M. A. Schembri (2000). "Fimbrial surface display systems in bacteria: from vaccines to random libraries." Microbiology 146 Pt 12: 3025-3032.
Konishi, E., S. Kosugi, et al. (2006). "Dengue tetravalent DNA vaccine inducing neutralizing antibody and anamnestic responses to four serotypes in mice." Vaccine 24(12): 2200-2207.
Korhonen, T. K., E. Tarkka, et al. (1983). "Type 3 fimbriae of
Klebsiella sp.: molecular characterization and role in bacterial adhesion to plant roots." J Bacteriol 155(2): 860-865.
Modis, Y., S. Ogata, et al. (2005). "Variable surface epitopes in the crystal structure of dengue virus type 3 envelope
glycoprotein." J Virol 79(2): 1223-1231.
Nassif, X. and P. J. Sansonetti (1986). "Correlation of the virulence of Klebsiella pneumoniae K1 and K2 with the presence of a plasmid encoding aerobactin." Infect Immun 54(3): 603-608.
28
Old, D. C. and R. A. Adegbola (1982). "Haemagglutinins and fimbriae of Morganella, Proteus and Providencia." J Med Microbiol 15(4): 551-564.
Old, D. C. and R. A. Adegbola (1985). "Antigenic relationships among type-3 fimbriae of Enterobacteriaceae revealed by immunoelectronmicroscopy." J Med Microbiol 20(1): 113-121. Old, D. C. and S. S. Scott (1981). "Hemagglutinins and fimbriae of
Providencia spp." J Bacteriol 146(1): 404-408.
Ong, C. L., G. C. Ulett, et al. (2008). "Identification of type 3 fimbriae in uropathogenic Escherichia coli reveals a role in biofilm formation." J Bacteriol 190(3): 1054-1063.
Regue, M., B. Hita, et al. (2004). "A gene, uge, is essential for
Klebsiella pneumoniae virulence." Infect Immun 72(1): 54-61. Reverchon, S., W. Nasser, et al. (1994). "pecS: a locus controlling
pectinase, cellulase and blue pigment production in Erwinia chrysanthemi." Mol Microbiol 11(6): 1127-1139.
Rosen, D. A., J. S. Pinkner, et al. (2008). "Utilization of an intracellular bacterial community pathway in Klebsiella
pneumoniae urinary tract infection and the effects of FimK on type 1 pilus expression." Infect Immun 76(7): 3337-3345. Sakellaris, H., D. P. Balding, et al. (1996). "Assembly proteins of
CS1 pili of enterotoxigenic Escherichia coli." Mol Microbiol
21(3): 529-541.
Sauer, F. G., M. Barnhart, et al. (2000). "Chaperone-assisted pilus assembly and bacterial attachment." Curr Opin Struct Biol
10(5): 548-556.
Schultz, J. E. and A. Matin (1991). "Molecular and functional characterization of a carbon starvation gene of Escherichia coli." J Mol Biol 218(1): 129-140.
Schwan, W. R., S. Shibata, et al. (2007). "The two-component response regulator RcsB regulates type 1 piliation in Escherichia coli." J Bacteriol 189(19): 7159-7163. Stout, V. and S. Gottesman (1990). "RcsB and RcsC: a
two-component regulator of capsule synthesis in Escherichia coli." J Bacteriol 172(2): 659-669.
Struve, C., M. Bojer, et al. (2009). "Identification of a conserved chromosomal region encoding Klebsiella pneumoniae type 1
29
and type 3 fimbriae and assessment of the role of fimbriae in pathogenicity." Infect Immun 77(11): 5016-5024.
Tarkkanen, A. M., B. L. Allen, et al. (1990). "Type V collagen as the target for type-3 fimbriae, enterobacterial adherence
organelles." Mol Microbiol 4(8): 1353-1361.
Valenski, M. L., S. L. Harris, et al. (2003). "The Product of the fimI gene is necessary for Escherichia coli type 1 pilus
biosynthesis." J Bacteriol 185(16): 5007-5011.
Yeh, K. M., A. Kurup, et al. (2007). "Capsular serotype K1 or K2, rather than magA and rmpA, is a major virulence determinant for Klebsiella pneumoniae liver abscess in Singapore and Taiwan." J Clin Microbiol 45(2): 466-471.
Zalewska, B., R. Piatek, et al. (2003). "Chimeric Dr fimbriae with a herpes simplex virus type 1 epitope as a model for a
30
Table 1. Bacteria strains used in this study
Strains Genotypes or relevant properties Reference or source
E. coli
JM109 RecA1 supE44 endA1 hsdR17 gyrA96 RelA1 thiΔ(lac-proAB)
Laboratory stock
S17-1 λ pir Tpr Smr recA, thi, pro,
hsdR-M+[RP4-2-Tc::Mu:KmrTn7](λ pir)
Laboratory stock
K. pneumoniae
CG43 K2 serotype Laboratory stock
CG43S3 ΔrspL, Str Laboratory stock
CG43S3-Z01 CG43S3 ΔlacZ Smr Laboratory stock
31
Table 2. Plasmids used and constructed in this study
Plasmids Relevant characteristic Reference or source
yT&A Cloning vector;Apr Yeastern Biotech
pGEMT Cloning vector;Apr Promega
pMrkABC mrkABC gene cluster cloned into pGEMT vector,Apr Laboratory stock pMrkABCD 1 kb fragment amplified using primer pairs, phw03 and MZ006,
and cloned into pMrkABC by AscI and ApaI site, Apr
Laboratory stock
pA25 5 amino acid insered in the 94th amini acid of MrkA of pMrkABCD
Laboratory stock
pD27 5 amino acid insered in the 87th amini acid of MrkA of pMrkABCD
Laboratory stock
pA-2 insert DENIIEDIII part2 in the pA25 PmeI site This study pD-1 insert DENIIEDIII part1 in the pD27 PmeI site This study pD-2 insert DENIIEDIII part2 in the pD27 PmeI site This study pD-3 insert DENIIEDIII part3 in the pD27 PmeI site This study pD-4 insert DENIIEDIII part4 in the pD27 PmeI site This study pD-5 insert DENIIEDIII part5 in the pD27 PmeI site This study
pLacZ15
A derivative of pYC016, containing a promoterless lacZ from K. pneumoniae CG43S3 as the reporter, Cmr
32
pMrkA-lacZ primers pmrkA4 and pmrkA5 were used to get 545 bp sequence, BamHI and BglII were used to cut into pLacZ15
33
Table 3. Primers used in this study
Primer Sequence 5'-->3' SL1138-3-1 GTTTAAACggaaaggaatgtcatactcta SL1138-3-2 GTTTAAACaaacacaacatggaacaatagtt SL1138-3-3 GTTTAAACagatcccttttgagataatggatttggaa SL1138-3-4 GTTTAAACcagtcaacccaatcgtaacagaa SL1138-3-5 GTTTAAACgagacagctacatcatcataggagta SL1139-3-1 GTTTAAACgtttctgctatttccttcacaactttaaactt SL1139-3-2 GTTTAAACatcttacatggagaaccgtccccttc SL1139-3-3 GTTTAAACacgactgtaatcaggcgacctaaaacatgtct SL1139-3-4 GTTTAAACtctccgaatggaggttctgcttctat SL1139-3-5 GTTTAAACactcctttcttaaaccagttgag
34
Table 4.DENIIEDIII inserted sequence and insertion site
Insertion site DENIIEDIII inserted sequence
A25-2 NMFKQTQHGTIVIRVQYEGDGSPCKMFKQNT D27-1 PVFRKGMSYSMCTG KFKVVKEIAETFKQPT D27-2 PVFKQTQHGTIVIRVQYEGDGSPCKMFKQPT D27-3 PVFKQIPFEIMDLEKRHVLGRLITLFKQPT D27-4 PVFKPVNPIVTEKDSPVNIEAE PPFGEFKQPT D27-5 PVFKRDSYIIIGVEPGQLKLNWFKKGMFKQPT
underline: DENIIEDIII amino acid
35
Fig. 1. Organization of the fimbriae encoding gene cluster in K. pneumoniae C3091 (http://www.ncbi.nlm.nih.gov/).
The mrkABCDF is coding for type 3 fimbriae. The gene cluster from fimB to fimK encode the regulatory system, for the expression and assembly, and structural components of type 1 fimbriae. The genes named pecM and pecS are homologs of the genes coding for the regulators for the expression of virulence genes in Erwinia chrysanthem (Reverchon, Nasser et al. 1994).
36
Fig. 2. Downstream genes of the type 3 fimbriae gene cluster. The genes KP4551, KP4552, and KP4554 were predicted
byNCBI ( http://www.ncbi.nlm.nih.gov/ ) to encode c-di-GMP binding domain protein (PilZ domain protein), transcription
37
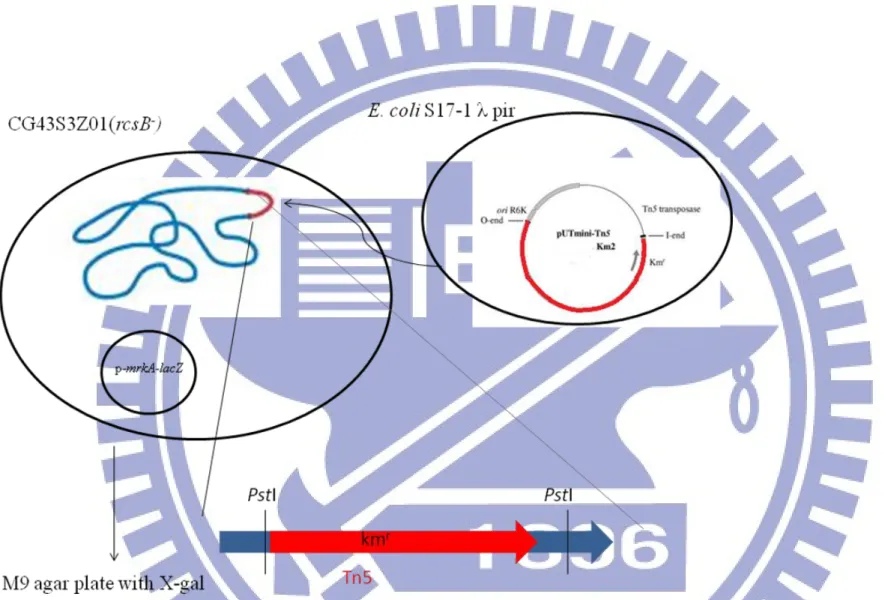
Fig. 3. Schematic presentation of the transposon mutagenesis. The plasmid pUTmini-Tn5Km2 was mobilized from E.
coli S17-1 λ pir to K. pneumoniae CG43S3Z01rcsB-, and the mutants selected on M9 agar plates containing kanamycin, chloramphinicol, and X-gal.
38
Fig. 4. Amino acid sequences of the major subunit of type 3 fimbriae. The arrows indicate A25 and D27 are the foreign
39
(A)
(B)
* : primer
Fig. 5. Amino acid sequences of DEN2EDIII. (A) Multiple-sequence
alignment of domain III region (aa 295 to 395) of four DV serotypes. Identical residues are shaded as dark gray areas, and conserved residues are shaded as light gray areas (Abd-Jamil, Cheah et al. 2008).The 5 fragmented amino acid sequences of are marked with different colors. (B) Sequences of the five peptides, the primers used for the PCR amplification are labeled.
40
(A)
(B)
41
Fig 6. Analysis of expression of the recombinant fimbriae. E. coli
JM109 carrying each of the recombinant clones were grown overnight at 37°C, and the lysates were collected and heated at 95°C for 30 mins before subjected to the analysis. (A) SDS-13.5% polyacrylamide gel stained with Coomassie blue. (B) and (C) are western blot analysis against anti-MrkA antibody and anti-DENEDIII antibody, respectively. The arrowheads mark the recombinant MrkA. Lanes M, molecular weight marker; 1, E. coli JM109; 2, JM109[pmrkABCD]; 3,
JM109[pD27]; 4, JM109[pA25-2]; 5, JM109[pD27-1]; 6, JM109[pD27-2]; 7, JM109[pD27-3]; 8, JM109[pD27-4]; 9, JM109[pD27-5].
42
(A)
(B)
Fig 7. Polymer pattern analysis of the recombinant fimbriae. E. coli
JM109 carrying each of the recombinant clones were grown overnight at 37°C and the total cell lysates collected by centrifugation. (A) SDS–8% polyacrylamide gel stained with Coomassie blue. (B) Western blot analysis by anti-MrkA antibody. Lanes M, Marker; 1, E. coli JM109; 2, JM109[pmrkABCD]; 3, JM109[pD27]; 4, JM109[pA25-2]; 5,
JM109[pD27-1]; 6, JM109[pD27-2]; 7, JM109[pD27-3]; 8, JM109[pD27-4]; 9, JM109[pD27-5].
43
(A)
(B)
Fig. 8. Immunofluorescence microscopy (IFM) analysis of the recombinant E. coli displaying with the type 3 fimbriae using the antibody (A)anti-MrkA and (B)anti-DENDIII. (a) JM109, (b) JM109
[pmrkABCD], (c) JM109[pD27], (d) JM109[pA25-2], (e) JM109[pD27-1], (f) JM109[pD27-2], (g) JM109[pD27-3], (h) JM109[pD27-4], (i) JM109[pD27-5].
44
(A)
(B)
Fig 9. Analysis of the purified fimbriae. The E. coli JM109 carrying
each of the recombinant plasmids were cultured for 24 h in 24 ml LB broth. The bacteria were collected by centrifugation for 3 min (8,000 rpm, 4°C) and were suspended in 1 ml phosphate-buffered saline (PBS). The bacterial suspension was heated at 65°C 2 h and homogenized in blender for 20 min at ambient temperature. 30% saturate ammonium sulfate was added and stored at 4°Covernight. The pellet was collected by centrifugation (20,000 g, 90 min) and suspended in 40 l PBS. The purified fimbriae were identified by (A) SDS–8% polyacrylamide gel stained with Coomassie blue. (B) Western blot analysis using anti-MrkA
45
antibody. Lanes M, Marker; 1, E. coli JM109; 2, JM109[pMrkABCD]; 3, JM109[pD27]; 4, JM109[pA25-2]; 5, JM109[pD27-1]; 6,
JM109[pD27-2]; 7, JM109[pD27-3]; 8, JM109[pD27-4]; 9, JM109[pD27-5].
46
(A)
(B)
Fig. 10. Analysis of the expression of the type 3 fimbriae in K.
pneumoniae CG43S3Z01 and K. pneumoniae CG43S3Z01rcsB-. The
bacteria (lanes 1 to 4) were cultured statically in LB for 20 h. The bacteria (lanes 6 to 9) were in shaking culture to OD 0.8. Total cell lysates were heated at 95°C for 30 min before subjected to analysis. (A) SDS–13.5% polyacrylamide gel stained with Coomassie blue;
(B)Western blot analysis using anti-MrkA antibody. The arrowheads mark the recombinant MrkA. Lanes M, Marker; 1 and 6,
47
CG43S3Z01[placZ]; 2 and 7, CG43S3Z01rcsB-(placZ);3 and 8,
CG43S3Z01(pmrkA-lacZ); 4 and 9, CG43S3Z01rcsB- (pmrkA-lacZ); 5, E. coli JM109.
48
(A)
(B)
49
Fig. 11. Analysis of the transposon insertion mutant strains. (A) The
mutant strains (M1~M8) with color alteration on X-gal plate are shown. (B) Analysis of the recombinant clone (in pUC19) by restriction enzyme PstI. Lanes 1: pM1; 2: pM2; 3: pM5; 4: pM6; 5: pM8; 6: pM7. (C) Analysis of the recombinant clone of pM1 (lanes 1, 3, 5) and pM6 (lanes 2, 4, 6) by restriction enzymes BamHI (lanes 1, 2), EcoRI (lanes 3, 4) and HindIII (lanes 5, 6).
50 (A) M2 (B) M5 (C) M7 (D) M8
51
Fig. 12. The tansposon insertion site and the genes beside the insertion site. The arrows mark the transposon insertion