溶藻弧菌胺醯組胺酸雙胜肽酶功能性胺基酸於催化特性分析之研究
全文
(2) 溶藻弧菌胺醯組胺酸雙胜肽酶功能性胺基酸於催化特性分析 之研究 Characterization of Functional Residues for the Catalysis of Aminoacylhistidine Dipeptidase from Vibrio alginolyticus 研究生:高良瑋. Student: Liang-Wei Kao. 指導教授:吳東昆 博士. Advisor: Dr. Tung-Kung Wu. 國 立 交 通 大 學 生物醫學研究所 碩士論文. A Thesis Submitted to Department of Biological Science and Technology College of Science National Chiao Tung University in partial Fulfillment of the Requirements for the Degree of Master in Biomedical Science August, 2008 Hsinchu, Taiwan, Republic of China. 中華民國九十七年八月.
(3) 溶藻弧菌胺醯組胺酸雙胜肽酶功能性胺基酸於催化特性分析 之研究 研究生:高良瑋. 指導教授:吳東昆 博士. 國立交通大學 生物醫學研究所碩士班. 摘要 實驗室從溶藻弧菌 ATCC 17749 中選殖出胺醯組胺酸雙胜肽酶(PepD)之基因並解 出序列。胺醯組胺酸雙胜肽酶(PepD, EC 3.4.13.3)為胜肽酶家族 M20 中的一員,可水解 雙胜肽 L-carnosine 及其他特定 Xaa-His 雙胜肽。此基因之開放讀碼區(ORF)序列共有 1473 個鹼基對,可轉譯出一條長 490 個胺基酸的蛋白質,計算其分子量約為 53.6 kDa, 並且此胺基酸序列和其他弧菌屬之 PepD 蛋白質序列比對有非常高之相似度。過去對於 細菌中胺醯組胺酸雙胜肽酶的研究很少,只針對其序列和部分生化特性進行探討,並無 其活性區胺基酸或金屬離子方面相關之研究。根據先前實驗室所架構的 pET-28a(+)-pepD 質體,表現出 N 端帶有 His-tag 之重組蛋白,並利用 Ni-NTA 親和層析 管柱純化。經由序列及結構分析預測溶藻弧菌 PepD 蛋白上胺基酸位置 His80、Asp82、 Asp119、Glu149、Glu150、Asp173、His219、Asn260、Arg369、Gly435 及 His461 為活 性或受質鍵結區域。將所預測影響金屬鍵結之胺基酸 Glu150 以及扮演固定 His80 使其 精確的與金屬鍵結的幕後推手 Asp82,分別進行定點突變;發現大部分突變蛋白皆失去 或降低原有之活性。此外,在預測受質結合位置 His219、Asn260、Arg369 及 Gly435 對其做丙氨酸突變試驗,並證明出 Arg369 為會影響活性之重要胺基酸。而 PepD 為含 金屬之蛋白質,金屬離子對其活性有直接的影響,而其中 EDTA 可抑制其活性主要原 因為失去活性區之金屬;以此為方向置換 PepD 活性中心金屬為其它二價金屬離子,明 確表示出此酵素對金屬的依賴,並且結果為含有不同金屬之 PepD 具有不同表現的活. I.
(4) 性。因此,根據本論文實驗結果將提出胺醯組胺酸雙胜肽酶活性區胺基酸之分佈情形與 金屬離子扮演重要之功能。. II.
(5) Characterization of Functional Residues for the Catalysis of Aminoacylhistidine Dipeptidase from Vibrio alginolyticus Student: Liang-Wei Kao. Advisor: Dr. Tung-Kung Wu. Institute of Biomedical Science National Chiao Tung University. Abstract. Earlier investigations in our laboratory, a newly defined aminoacylhistidine dipeptidase from Vibrio alginolyticus ATCC 17749 was characterized via the determination of the corresponding gene sequence. Aminoacylhistidine dipeptidase (PepD, EC 3.4.13.3) is a member of the peptidase family, which catalyzes the cleavage and release of N-terminal amino acid from a dipeptide molecule like L-carnosine (β-Ala-L-His). The pepD gene from Vibrio alginolyticus encodes a polypeptide of 490 amino acids, which has a sequence that is highly similar to that of dipeptidases from various other species. The researches on bacterial PepD were less known and only investigated genetically and biochemically. Previously, the gene was cloned into the pET-28a(+) expression vector and expressed as a (His)6-PepD fusion protein, which was purified via a Ni-NTA column. The sequence alignment and structure model of PepD indicate that His80, Asp119, Glu150, Asp173 and His461 are metal binding site residues, and Asp82, Glu149, His219, Asn260, Arg369, and Gly435 are putative catalysis and substrate binding residues. Site-directed mutations of Asp82, Glu150 and Arg369 residues of PepD exhibit a decline or loss of activity, suggesting that these residues might be involved in substrate and metal binding, thereby dramatically affecting enzymatic activity. The native Zn2+ ion was removed from PepD using an EDTA chelating solution, which resulted in the loss of hydrolysis activity. However, PepD activity could be restored by III.
(6) adding Mg2+, Mn2+, Co2+, Ni2+, Cu2+ or Cd2+, indicating the functional importance of the metal ion. The functional role of these residues in enzyme catalysis and the effect of metal ions will be discussed in this thesis.. IV.
(7) 謝誌 (Acknowledgement) 在這兩年碩班的過程可以說是有苦有樂、有喜有悲,並且更加豐富了人生旅途的色 彩,最重要感謝的是我指導教授吳東昆 博士,他讓我感覺到只要有對的研究態度及認 真學習的熱誠,哪怕是不同背景的學生,並且在實驗挫折及茫然時教授都以適時的鼓勵 與關心取代責備,使得在研究所成長了非常多,就算成果失敗也要勇敢積極面對及不逃 避。以及感謝口試委員李耀坤 教授、袁俊傑 教授與鄭建中 教授於百忙之中抽空審閱 及修改我的論文初稿,並對實驗觀念、過程、結果與討論提供諸多寶貴意見,使本論文 能更加完善。 同時也非常感謝實驗室的所有成員:裕國,你是我的開山始主,感謝你;媛婷、晉 源,你們對我太好了,一切盡在不言中;小紅、晉豪,你們太可愛了;小麵包、天昶, 感謝你們讓我問怪問題及耐著性子回答,並且我們擁有許的話題;以及其他沒點到名 的,相同的也都非常感謝你們,很八股的對你們說:「沒有你們就沒有現在的小高。」 壓軸要感謝的是我的家人及朋友;首先是家人,爸媽感謝你們讓我在金錢方面完全 沒有負擔,並且無條件的讓我抱怨讓我依靠,哥姐感謝你們支持我讀書,並且於過程中 無限的提供資源,我愛你們。朋友方面先感謝那兩個損友兼室友,小帆帆及阿旻,讓我 每天回家後有人可以碎碎念並且給予建議;以及那群在新竹的玩咖,阿偉、小喇、阿蛇、 Lance、小班、萱萱、小米、小龐………還有好多人,謝謝讓我生活中除了 70%為實驗 外,另為 30%就是你們的了。另外分散各的好麻吉,怡仁、Tracy、流氓、小君……一 樣還是好多人,感謝你們陪我一路碰碰撞撞,並三不五時的慰問及約出去瘋。 最後,感謝現在正在觀看此論文的人,希望不成材的小弟對您可以有所幫助,並祝 各位一切順利、身體健康。. V.
(8) Keywords Vibrio alginolyticus, aminoacylhistidine dipeptidase, PepD, peptidase family M20, L-carnosine, metallopeptidase, site-directed mutagenesis. VI.
(9) Abbreviations APS. Ammonium persulfate. BCS. bovine calf serum. bp. base pair. BSA. bovine serum albumin. CD. Circular dichroism. CDC. Centers for Disease Control and Prevention. CO2. carbon dioxide. dH2O. distilled water. ddH2O. double distilled water. DMEM. Dulbecco’s Modified Eagle Medium. DMF. dimethylformamide. DMSO. dimethyl sulfoxide. DNA. deoxyribonucleic acid. dNTP. deoxynucleoside triphosphate. EDTA. ethylenediamine-tetraacetic acid. ELISA. enzyme-linked immunosorbent assay. FBS. fetal bovine serum. GABA-His. γ-Amino-butyryl-histidine dipeptide. HCl. hydrogen chloride. HRP. horseradish peroxidase. H2SO4. sulferic acid. ICP-MS. inductively coupled plasma-mass spectrometry. i.p.. intraperitoneal. IPTG. isopropyl-1-thio-β-D-galactopyranoside VII.
(10) i.v.. intravenous. kb. kilobase(s). kDa. kiloDalton(s). KPi. potassium phosphate. LB. Luria-Bertani. mAb. monoclonal antibody. NaCl. sodium chloride. NaN3. sodium azide. NaOH. sodium hydroxide. NC. nitrocellulose. NCBI. National Center for Biotechnology Information. NC-IUBMB. Nomenclature Committee of the International Union of Biochemistry and Molecular Biology. NEM. N-ethylmaleimide. OPA. o-phthaldialdehyde. pAb. polyclonal antibody. PBS. phosphate buffered saline. PCR. polymerase chain reaction. PEG. polyethylene glycol. pI. isoelectric point. RT. room temperature. s.c.. subcutaneous. SDS-PAGE. sodium dodecyl sulfate polyacrylamide gel electrophoresis. SEM. Scanning Electron Microscopy. spp.. species. VIII.
(11) TCA. trichloroacetic acid. TEMED. N,N,N’,N’-tetramethylethylenediamine. TMB. 3,3’,5,5’-tetramethylbenzidine. Tris base. Tris(hydroxymethyl)aminomethane. TSB. tryptic soy broth. X-gal. 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside. IX.
(12) Table of Contents Abstract (Chinese) ............................................................................................... I Abstract (English) ............................................................................................. III Acknowledgement ...............................................................................................V Keywords ........................................................................................................... VI Abbreviations .................................................................................................. VII Table of Contents ................................................................................................X List of Figures .................................................................................................. XII List of Tables ................................................................................................... XIV Chapter 1 Introduction....................................................................................... 1 1.1 Vibrio species and Vibrio alginolyticus ......................................................................... 1 1.2 Binuclear Metallohydrolase.......................................................................................... 3 1.3 Metallopeptidase and Metalloaminopeptidase ........................................................... 4 1.4 Peptidase family M20 .................................................................................................... 6 1.5 Aminoacylhistidine dipeptidase ................................................................................... 7 1.6 Other Carnosine-Hydrolyzing Enzymes ..................................................................... 9 1.6.1 Carnosinase ............................................................................................................ 10 1.6.2 Peptidase V ............................................................................................................. 12 1.7 Motivation and Research Objectives ......................................................................... 15 . Chapter 2 Methods ........................................................................................... 16 2.1 Expression of the V. alginolyticus pepD gene in E. coli ............................................. 16 2.2 Purification of the expressed Vibrio alginolyticus PepD........................................... 16 2.3 Protein concentration determination......................................................................... 17 2.4 SDS-PAGE and Native-PAGE analysis ..................................................................... 18 2.5 Enzymatic activity assay of PepD .............................................................................. 19 2.6 Mutagenesis analysis of V. alginolyticus pepD ........................................................... 20 X.
(13) 2.7 The His-tag-cleaved PepD .......................................................................................... 21 2.8 Metal ion effect of PepD activity ................................................................................ 22 2.9 Enzyme kinetics ........................................................................................................... 22 . Chapter 3 Results .............................................................................................. 24 3.1 Expression and Purification of Vibrio alginolyticus PepD ....................................... 24 3.2 Multiple sequence alignment and structural model ................................................. 25 3.3 Site-directed Mutagenesis Analysis of Vibrio alginolyticus PepD............................ 29 3.4 Metal ion effect of PepD activity ................................................................................ 34 3.5 Enzyme kinetics of the metal effects on Vibrio alginolyticus PepD ......................... 36 . Chapter 4 Discussion and Conclusions ........................................................... 38 Chapter 5 Future work ..................................................................................... 42 Chapter 6 Reference ......................................................................................... 43 Appendix 1 ......................................................................................................... 47 Appendix 2 ......................................................................................................... 48 Appendix 3 ......................................................................................................... 49 Appendix 4 ......................................................................................................... 50 Appendix 5 ......................................................................................................... 51 Appendix 6 ......................................................................................................... 52 Appendix 7 ......................................................................................................... 53 . XI.
(14) List of Figures Fig. 1:Binuclear metallohydrolases that catalyze the hydrolysis of peptide and phosphodiester bonds.. ................................................................................................ 3 Fig. 2: A generalized mechanistic scheme for metalloaminopeptidases.. ........................... 4 Fig. 3: An enzymatic reaction catalyzed by aminoacylhistidine dipeptidase. .................... 7 Fig. 4: (A) Ribbon diagram of the crystalline structure of PepV; and (B) Local view of the zinc-binding residues of PepV. ........................................................................... 13 Fig. 5: Schematic of the active site of PepV. ........................................................................ 14 Fig. 6: Formation of a Schiff base by L-histidine and o-phthalaldehyde ......................... 20 Fig. 7: The overall structure of PepD and the local view of catalytic and zinc binding residues (by Chin-Yuan Chang). .............................................................................. 25 Fig. 8: Local view of PepD (green) superimposed upon the substrate binding site of PepV (blue). ................................................................................................................ 26 Fig. 9: Nucleotide and predicted amino acid sequences of the V. alginolyticus pepD gene ..................................................................................................................................... 28 Fig. 10: SDS-PAGE (12%) of purified wild-type and mutant proteins of Asp82 and Glu150. ........................................................................................................................ 29 Fig. 11: Enzymatic activities of wild-type and mutant Asp82 and Glu150 PepD on L-carnosine ................................................................................................................. 30 Fig. 12: SDS-PAGE (12%) of purified wild-type and mutant proteins of His219, Asn260, Arg369, and Gly435. .................................................................................................. 31 Fig. 13: Enzymatic activities of wild-type and mutant His219, Asn260, Arg369, and Gly435 PepD on L-carnosine. ................................................................................... 32 Fig. 14: Enzyme kinetics of the mutant His219, Asn260, and Gly435 PepD proteins ..... 32 . XII.
(15) Fig. 15: Metal ion effect on PepD activity............................................................................ 34 Fig. 16: Enzyme kinetics of the metal effects on Vibrio alginolyticus PepD ..................... 36 Fig. 17: Proposed mechanism for the hydrolysis of PepD ................................................. 39 . XIII.
(16) List of Tables Table 1: Solutions and volumes for the preparation of the SDS-PAGE and Native-PAGE separating gel and stacking gel .............................................................................. 19 Table 2: Reaction conditions and cycling parameters for the PCR mutagenesis reaction ................................................................................................................................... 21 Table 3: The fluent gradient program ................................................................................. 22 Table 4: Kinetic parameters for the hydrolysis of L-carnosine using mutant V.alginolyticus PepD. ............................................................................................... 33 Table 5: Kinetic parameters for the hydrolysis of L-carnosine using different metal ion derivatives that complex with V. alginolyticus PepD. ........................................... 37 . XIV.
(17) Chapter 1 Introduction 1.1 Vibrio species and Vibrio alginolyticus Vibrio species (Vibrio spp.) are considered opportunistic pathogens in both aquaculture species and humans. Vibriosis is the major disease caused by Vibrio spp. in shrimp aquaculture, resulting in high mortality and severe economic loss in all producing countries. On the basis of phenotypic data, the major species causing vibriosis in shrimp are V. alginolyticus, V. anguillarum, V. harveyi, and V. parahaemolyticus[1]. V. anguillarum, V. damsela, and V. carchariae are major vibriosis-causing species in fish.. Oceanomonas alginolytica first was recognized and named by Miyamoto et al.[2]. It was renamed V. alginolyticus by Sakazaki in 1968.[3] V. alginolyticus is very similar to V. parahaemolyticus, in terms of their biochemical properties; they also are isolated from similar types of marine sample. Acetoin production and the fermentation of arabinose are important features which distinguish V. alginolyticus from V. parahaemolyticus.[4] V. alginolyticus has been found to have a worldwide distribution, including coastal waters, sediments, and seafood taken from temperate and tropical areas.[4] As with other Vibrio spp., V. alginolyticus appears on a seasonal basis, being rarely found in winter and quite abundant in summer. Studies by Baross and Liston have shown that the minimum growth temperature for V. alginolyticus is 8℃[5].. Grouper culture has become an alternative, important activity since the penaeid shrimp culture industry collapsed in the late 1980’s in Taiwan. The production of farmed grouper has doubled in Taiwan, from 5052 tonnes in 2001 to 12,103 tonnes in 2004. Vibriosis caused by pathogenic V. alginolyticus is a common problem in the intensive culture of grouper, causing. 1.
(18) a gastroenteritis syndrome (swollen intestines containing yellow fluid) [6]. In addition to its importance in the grouper culture, V. alginolyticus also is considered important in fish and shrimp culture pathogenicity, on account of previous data from field studies [7] and a disease outbreak within shrimp farming in 1996 [8].. V. alginolyticus first was recognized as a human pathogen in 1973 [9]. In recent years, several studies have demonstrated clinical infection caused by V. alginolyticus [10-12], an organisms that poses a risk to humans who are in contact with it. Most human infections caused by V. alginolyticus can be ascribed to the consumption of raw or undercooked seafood, including fish, shellfish, shrimps, and squid [13]. As with other Vibrio spp., the major clinical manifestations of illness with V. alginolyticus are gastroenteritis, wound infections, and septicemia. Wound infections through waterborne transmission are the most frequent V. alginolyticus infections, and account for 71% of all infections [14]. Gastroenteritis has been a relatively uncommon presentation of V. alginolyticus infection, but still accounts for 12% of infections [15]. Other reported manifestations of disease are ear infections, central nervous system disease and osteomyelitis [16,17].. 2.
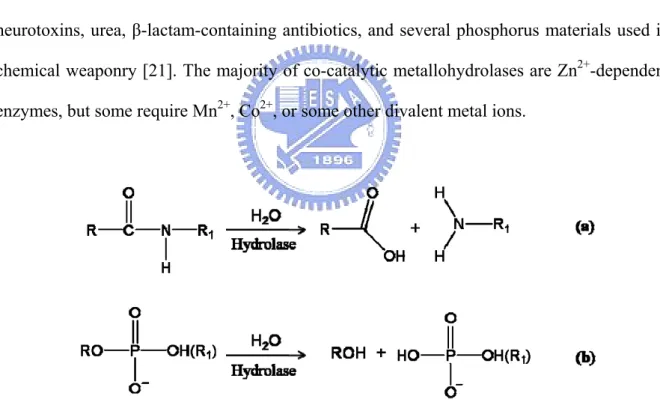
(19) 1.2 Binuclear Metallohydrolase Within metabolic and signaling biochemical pathways, there are numerous steps that involve the hydrolytic cleavage of peptide or phosphate ester bonds. Although both types of bonds are thermodynamically unstable to hydrolysis, there are significant kinetic barriers to these reactions [18]. Consequently, several hydrolases that contain co-catalytic metallo-active sites catalyze diverse reactions, such as the degradation of DNA, RNA, phospholipids, and polypeptides (Fig. 1). They are key players in carcinogenesis, tissue repair, protein maturation, hormone level regulation, cell-cycle control, and protein degradation [18-21]. In addition, co-catalytic metallohydrolases are involved in the degradation of agricultural neurotoxins, urea, β-lactam-containing antibiotics, and several phosphorus materials used in chemical weaponry [21]. The majority of co-catalytic metallohydrolases are Zn2+-dependent enzymes, but some require Mn2+, Co2+, or some other divalent metal ions.. Fig. 1:Binuclear metallohydrolases that catalyze the hydrolysis of peptide and phosphodiester bonds. (a) Peptidases that catalyze the hydrolysis of peptide bonds in polypeptides. (b) Phosphatases and nucleases that catalyze the hydrolysis of phosphodiester bonds in phosphorylated amino acids and saccharides, nucleotides, DNA, and RNA.. 3.
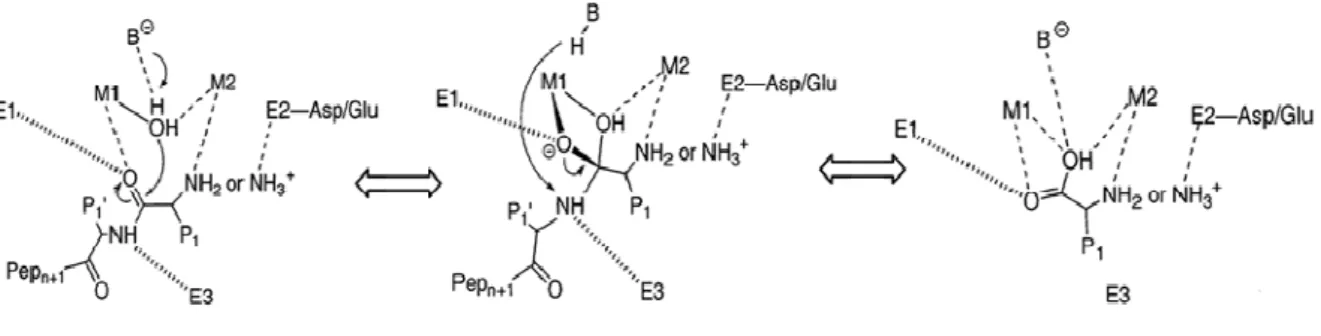
(20) 1.3 Metallopeptidase and Metalloaminopeptidase The class of peptidases that require a metal ion for their catalytic activity has been named metallopeptidases. Metallopeptidases can be divided into two broad types, depending upon the number of metal ions required for catalysis. In many metallopeptidases, only one metal ion is required; but, in some families, two metal ions must act together, or so called ‘co-catalytically’. All known co-catalytic metallopeptidases are exopeptidases, which include aminopeptidases,. carboxypeptidases,. dipeptidases,. and. tripeptidases;. meanwhile,. metallopeptidases with only one catalytic metal ion generally are exopeptidases or endopeptidases.. Metalloaminopeptidases represent a family of enzymes that use one or two metal ions to specifically cleave the N-terminal amino acid residues of polypeptides and proteins. They play fundamental roles in different biochemical events, such as protein maturation and degradation, tissue repair, and cell-cycle control [22]. In these enzymes, the nucleophilic attack on a peptide bond is mediated by a water molecule and the water molecule is activated by a divalent metal cation, usually zinc but sometimes cobalt, manganese, nickel or copper [23].. Fig. 2: A generalized mechanistic scheme for metalloaminopeptidases. First, the substrate binds to the active site, with the carbonyl group of the scissile peptide bond interacting with M1 and a conserved enzyme residue, E1. The N-terminus either interacts 4.
(21) with M2 or with one or more acidic enzyme residues in the vicinity, indicated in Figure 2 by E2. Additional enzymic histidine or backbone carbonyl group interactions, at E3, facilitate substrate binding in the correct register. The scissile peptide bond is attacked by a solvent molecule that has been activated by its interaction with the metal ion(s), and by an enzyme residue that functions as a general base, B. Whether or not the subsequent tetrahedral intermediate is stabilized by interactions to both metal ions and E2 side chains depends upon the particular enzyme system. Breakdown of the intermediate is most likely promoted by the addition of a proton to the amine group that departs from the former general base, B-H, as first suggested for thermolysin [23].. 5.
(22) 1.4 Peptidase family M20 The aminoacylase-1/metallopeptidase 20 (Acy1/M20) family, which is part of the metallopeptidase H (MH) clan, represents the largest group of metallopeptidases, based on sequence similarity. The MEROPS database of peptidases (http://merops.sanger.ac.uk) [24] is a record of over 1000 sequences for this family, derived from all kingdoms of life. According to the MEROPS database, metallopeptidases can be classified into 15 clans: MA, MC, MD, ME, MF, MG, MH, MJ, MK, MM, MN, MO, MP, MQ, and M-. The peptidases in clan MH contain a variety of co-catalytic zinc-dependent peptidases that bind two ions of zinc per monomer, which is held by five amino acid ligands[25] and inhibited by the general metal chelator, ethylenediamine-tetraacetic acid (EDTA).. The peptidase clan MH is further classified into four families: M18, M20, M28, and M42. Proteins of peptidase family M20 have been characterized as water bound by two zinc ions ligated by five residues, in the order His/Asp, Asp, Glu, Glu/Asp, and His at the active site. Additional Asp and Glu residues also are thought to be important for catalysis and to occur adjacent to metal-binding residues[26]. The Asp residue between two catalytic residues binds both metal ions. Several available crystal structures for M20 family enzymes, including PepV, CPG2, exhibit a dizinc-binding domain. Enzymes of the Acyl/M20 family have demonstrated the potential for a variety of applications. However, variations exist in the individual subfamilies that can be seen in their amino acid sequence alignments. Peptidase family M20 can be devided into 4 subfamilies - M20A, M20B, M20C, and M20D - with the active site residues different among the subfamilies.. 6.
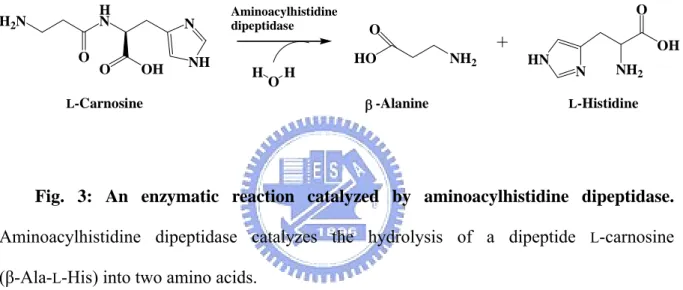
(23) 1.5 Aminoacylhistidine dipeptidase According to the peptidase family database, aminoacylhistidine dipeptidase (EC 3.4.13.3, also Xaa-His dipeptidase, X-His dipeptidase, carnosinase, and PepD) is assigned to the M20C subfamily, which are zinc-containing metallopeptidases, and which catalyze the cleavage and release of an N-terminal amino acid, usually a neutral or hydrophobic residue, from Xaa-His dipeptides or polypeptides (Fig. 3)[26].. H2 N O. H N. N. O. NH. OH. Aminoacylhistidine dipeptidase. O O HO. H. O. H β -Alanine. L-Carnosine. NH2. OH HN. N. NH2. L-Histidine. Fig. 3: An enzymatic reaction catalyzed by aminoacylhistidine dipeptidase. Aminoacylhistidine dipeptidase catalyzes the hydrolysis of a dipeptide L-carnosine (β-Ala-L-His) into two amino acids.. This gene exists extensively in prokaryotes and eukaryotes. In 1974, the first direct proof of aminoacylhistidine dipeptidase activity in the hydrolysis of an unusual dipeptide L-carnosine (β-Ala-L-His) in bacteria was obtained in the organism Pseudomonas aeruginosa. [27], and studies suggested that the expression of pepD negatively affects biofilm formation, which is considered necessary for infection, thereby causing fish mortality and significant economic loss [28]. Therefore, PepD could be a promising target to control bacterial biofilm formation and infection. In subsequent years, this carnosine-hydrolyzing enzyme has been identified in a number of bacterial species, but only the PepD of Escherichia coli has been characterized genetically and biochemically [29]. The pepD from E. coli encodes a 52 kDa. 7.
(24) protein and is active as a homodimer, with a molecular mass of 100 kDa[30]. The pure enzyme exhibits a pH and temperature optimum of pH 9.0 and 37℃, respectively. E. coli PepD appears to be a metallopeptidase with broad substrate specificity, which is activated by Co2+ and Zn2+, and deactivated by metal chelators [29]. In general, dipeptidases are involved in the final breakdown of the protein degration fragments produced by other peptidases, or in the final dipeptide breakdown for amino acid utilization. The same result has been observed for PepD-deficient mutants of E. coli[31] and S. typhimurium[32], which indicates that PepD hydrolyzes dipeptide as an amino acid source. However, the biological impact of PepD remains unclear.. 8.
(25) 1.6 Other Carnosine-Hydrolyzing Enzymes Several other proteins have been reported to exhibit dipeptidase activity on L-carnosine. In mammals, at least two types of carnosinase exist with different properties (see 1.6.1). The first enzyme is a serum carnosinase, and the second one is known as a cytosolic form [33]. Peptidase V from Lactobacillus delbrueckii ssp. originally was identified as a carnosinase, cleaving L-carnosine as a source of histidine (see 1.6.2)[34]. BapA from Pseudomonas sp., proposed as a β-Ala-Xaa dipeptidase (EC 3.4.13.-), has been found to hydrolyze the peptide bonds of β-alanyl dipeptides (β-Ala-Xaa) [35]. Pep581 from Prevotella species (Prevotella spp.) shares 47% sequence identity with E. coli PepD, and has a calculated molecular weight of 53.2 kDa. Pep581 hydrolyzes both dipeptides and a single amino acid from the N-terminus of tri- and oligopeptides, so that it differs from PepD enzymes [36].. 9.
(26) 1.6.1 Carnosinase In mammals, at least two types of carnosinase exist with different properties. The first enzyme is serum carnosinase (CN1, EC 3.4.13.20)[33] and the second one is cytosolic carnosinase (also named tissue carnosinase, CN2, EC 3.4.13.18). Serum carnosinase (CN1) has been identified as a homodimeric dipeptidase with restricted substrate specificity for Xaa-His dipeptides, including L-carnosine. Carnosine is synthesized in many tissues by carnosine synthase (EC 6.3.2.11) from β-alanine and histidine, and is degraded by intra- or extracellular dipeptidases, also named carnosinases, all belonging to the large family of metalloproteases. The nature of the metal ion in serum carnosinase remains unknown, and could be activiated by Cd2+ and citrate ions[37]. It also could hydrolyze homocarnosine and anserine, activities that are not inhibited by bestatin, a compound known to specifically inhibit various amino- and di-peptidases.. Serum carnosinase has been assumed to be involved in certain important pathological conditions. Decreased concentrations of serum carnosinase have been detected in patients with Parkinson’s disease and multiple sclerosis, and after a cerebrovascular accident [38]. It also has been suggested that monitoring serum carnosinase might be useful to predict the clinical course of patients after an acute stroke [39]. Deficiency of human carnosinase has been associated with neurological deficits, including intermittent seizures and mental retardation [40,41]. Studies on serum carnosinase also have been approached by means of computational analysis, and have suggested some therapeutic usefulness of either inhibition by L-carnosine analogues (e.g., in diabetes) or activation of the enzyme via the rational design of citrate-like, non-toxic allosteric modulators (e.g., in homocarnosinosis) [42].. Tissue carnosinase (CN2) first was isolated from porcine kidney by Hanson and Smith. 10.
(27) in 1949[43] and subsequently found to be widely distributed within the tissues of rodents and higher mammals. It acts as a ubiquitous nonspecific dipeptidase, rather than as a selective carnosinase with broad substrate specificity, but does not hydrolyze homocarnosine or anserine [33]. This enzyme requires Mn2+ ions for its activity and is strongly inhibited by low concentrations of bestatin.. 11.
(28) 1.6.2 Peptidase V Peptidase V (PepV), from Lactobacillus delbrueckii ssp. lactis DSM 7290, originally was identified as a carnosinase, cleaving L-carnosine as a source of histidine, in the E. coli mutant strain UK197 (pepD, hisG) [44]. Since then, it also has been characterized as a relatively non-specific dipeptidase, cleaving a variety of dipeptides, especially those with an unusual β-alanyl residue in the N-terminus, and removing the N-terminal amino acid from a few distinct tripeptides [44]. Moreover, PepV is related not only to peptidases, but also to acetylornithine deacetylase (ArgE, EC 3.5.1.16) and succinyldiaminopimelate desuccinylase (DapE, EC 3.5.1.16), and recently has been described as a member of the aminoacylase-1 family [45]. These enzymes share the characteristics of hydrolyzing amide bonds in a zinc(or cobalt-) dependent manner. Therefore, PepV is recognized as a metallopeptidase in the M20A subfamily from the MH clan. It can be fully inhibited using metal chelating agent 1 (MCA-1), 10-phenanthroline or EDTA.. The Lactobacillus delbrueckii PepV protein was the first discovered crystallized dinuclear dipeptidase with carnosine-hydrolyzing enzymatic activity in the M20 family. The Lactobacillus delbrueckii pepV gene is 1413 nucleotides in length, consists of 470 amino acid residues, and encodes a protein with a predicted molecular mass of 52 kDa. Lactobacilli are organisms with multiple amino acid auxotrophies, making them critically dependent upon their proteolytic abilities to efficiently degrade milk protein casein. Consequently, they are used as starting materials during dairy fermentation. Deletion of the dipeptidase pepV gene from Lactobacilli results in significantly decreased growth rates, but does not reduce final cell density [34].. The 3D structure of PepV protein consists of two distinct domains, named the lid (lower). 12.
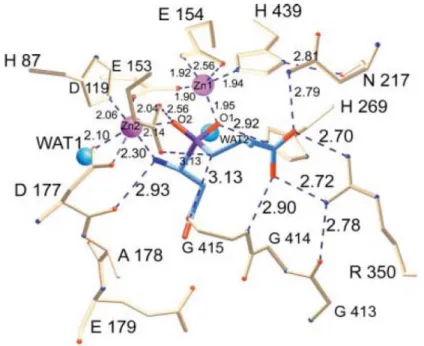
(29) domain and the catalytic (upper) domain (Fig. 4A). The upper domain contains the polypeptides from Met1 to Gly185 and from Ser388 to Glu468, whereas the lid domain is comprised of residues Glu186 to Gly387. In the crystalline structure of PepV, two zinc ions are associated in one monomer protein [46]. Zinc ions are located in the catalytic domain and held by five residues, including His87, Asp119, Glu154, Asp177 and His439 (Fig. 4B).. Fig. 4: (A) Ribbon diagram of the crystalline structure of PepV; and (B) Local view of the zinc-binding residues of PepV. The inhibitor of PepV (beige) is superimposed upon the zinc binding residues of AAP (blue) and CPG2 (red). Residues are numbered according to PepV sequence. The catalytic water molecule of CPG2 is depicted in red (WAT).. It is notable that the peptide group between the bridging Asp119 and the adjacent residue Asp120 exhibits a cis-conformation. Furthermore, the imidazole Nδ1 atoms of His439 (zinc 1) and His87 (zinc 2) are further clamped to Asn217 O and Asp89 O, respectively. These two zinc ions, as described by Jozic et al. [46], are considered to play two different roles in the hydrolysis of substrates: in the stabilization of the substrate-enzyme 13.
(30) tetrahedral intermediate; and in the activation of the catalytic water molecule (Fig. 5). Zinc 1, which is associated with the imidazole group of H439, and the carboxylate oxygens of E154 and D119 primarily appear to facilitate substrate binding via an ‘oxyanion binding hole’, with H269 resulting in polarization of the scissile carbonyl group, thereby promoting nucleophilic attack by the catalytic water molecule. Zinc 2 is coordinated by the carboxylate oxygens of H87 and D177 and of the bridging D119. It seems primarily to activate the catalytic water molecule and to promote binding and hydrolysis.. Fig. 5: Schematic of the active site of PepV. The Asp-Ala phosphinate inhibitor mimics the dipeptide substrate, as shown in blue. The bridging catalytic water attacks the carbonyl carbon of the scissile peptide bond to form an sp3-orbital substrate-enzyme tetrahedral intermediate.. 14.
(31) 1.7 Motivation and Research Objectives Aminoacyl-histidine dipeptidase (PepD) exists extensively among prokaryotes and eukaryotes, and belongs to the metallopeptidase 20 (M20) family within the metallopeptidase H (MH) clan (MEROPS: the peptidase database). Enzymes of the peptidase M20 family have shown potential for different applications, like being an anti-bacterial target and as a therapeutic agent for cancer treatment. They also may play a role in combating aging and neurodegenerative or psychiatric diseases. Several metallopeptidases that contain co-catalytic metallo-active sites are key players in cell-cycle control, tissue repair, and protein degradation. The majority of co-catalytic metallohydrolases are Zn2+-dependent enzymes, but some require Mn2+, Co2+, or some other divalent metal ion. Interestingly, these metal ions play an important role in these kinds of enzyme. However, the sequence alignment of PepD suggests that five residues - His80, Asp119, Glu150, Asp173 and His461 - hold the catalytic metal ions. Furthermore, the structure model indicates that Asp82, Glu149, H219, N260, R369 and G435 are located in an important position. In order to understand these important characterizations of PepD, a study of the functional residues and metal ion effect was undertaken.. 15.
(32) Chapter 2 Methods. 2.1 Expression of the V. alginolyticus pepD gene in E. coli We used pET-28a(+) as the expression vector. Constructed pET-28a(+)-pepD plasmids were transformed into E. coli BL21(DE3)pLysS competent cells by means of the heatshock method, and the cells spread on an LBkan agarose plate and incubated at 37ºC for 12 to 16 hrs. Colonies harboring pET-28a(+)-pepD were extracted and cultured in 3 ml LBkan medium for several hours, and then transferred into a 300 mL LBkan medium. 150 μL 1 M IPTG (final concentration was 0.5 mM) was added when the OD600 approached 0.5 ~ 0.6, and then the colonies incubated at 37ºC for another 6 hrs to induce the expression of PepD protein. pET-28a(+) plasmids also were transformed into E. coli BL21(DE3)pLysS competent cells, following the same experimental procedure as for controls.. 2.2 Purification of the expressed Vibrio alginolyticus PepD After 6 hrs incubation at 37ºC, with rotary shaking, cells were collected by centrifugation at 6,500rpm for 30 min at 4ºC. The bacterial pallet was resuspended with 20 mL 20 mM Tris-HCl, 0.5 M NaCl, pH 6.8 buffer (buffer A). The resuspended cells were disrupted by sonication using a sonicator pulsing in a 2 sec on, 1 sec off pulsation cycle for the total sonication time of 3 min at 30% energy. The entire experimental sonication procedure was conducted on ice. The sonication steps were repeated at least 2 additional times. After sonication, the cell lysate was centrifuged at 9,500rpm for 30 min at 4ºC to remove the cell debris and intact cells. The supernatant was collected for further purification.. 16.
(33) The supernatant was purified by affinity chromatography using a Ni-NTA column. One mL Ni-NTA resin was packed in a 20 mL plastic column and pre-equilibrated with 10 mL buffer A containing 20 mM imidazole (10 bed volume). The supernatant was loaded into the column and then washed with 10 mL buffer A containing 20 mM imidazole (5 bed volumes). Five bed volumes of buffer A, consisting of 80 mM, 150 mM, 300 mM, or 500 mM imidazole, were used sequentially to elute the expressed PepD protein. Finally, the Ni-NTA column was washed with buffer A, containing 1 M imidazole. The eluted fractions were collected for SDS-PAGE analysis and enzymatic activity assay. By SDS-PAGE analysis, the high-purity eluted fractions were collected and dialyzed with 2 L 50 mM Tris-HCl pH 6.8 buffer for 2 hrs, followed by 3 L for 8 hrs. After enzymatic activity analysis, the purified proteins were stored at -80ºC to be ready for subsequent experiments. Note that PepD can be stored at -80 ºC for six months with no loss of of activity.. 2.3 Protein concentration determination The protein concentrations of purified proteins were measured using BCA Protein Assay Reagents. To each well of the F96 MicroWellTM plate was added a 20 μL sample mixed with 200 μL BCATM Working Reagents (BCATM Reagent A:BCATM Reagent B = 50:1). The reactions were incubated at 37ºC for 30 min in the dark. The absorbances of samples were measured at 562 nm on a Multiskan Ascent Microplate Reader. 2 mg/mL bovine serum albumin (BSA) stock and successive dilutions (1.5, 1.0, 0.75, 0.5, 0.25, 0.125, 0.025 mg/mL) served as standards, following the same procedure described above.. 17.
(34) 2.4 SDS-PAGE and Native-PAGE analysis After expression and purification, gel electrophoresis was used to assess for protein expression level, purity, and molecular weight. The samples were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS PAGE) on 12.5% gels (Table 2). Each 10 μL sample was mixed with 2 μL 5X SDS-PAGE sample buffer and incubated at 95ºC for 5 min to denature proteins. Electrophoresis was performed with 1X SDS-PAGE running buffer at 90 Volts for 30 min, followed by 120 Volts for 1.5 hrs. The SDS-PAGE gel was stained with a stain buffer containing Coomassie Brilliant blue R-250 for 30 min and destained with destain buffer I (methanol/acetic acid/water = 4:1:5, v/v/v) for 20 min, followed by destain buffer II (methanol/acetic acid/water = 1.2:0.05:8.75) overnight.. Native-PAGE was performed to examine the native form of PepD. The purified and dialyzed protein fractions were separated by Native-PAGE on 7.5% gels (Table. 1). The experimental steps were similar to SDS-PAGE analysis, except that the gel contained no SDS and there was no denaturing treatment. Each 10 μL sample was mixed with 2 μL 5X Native-PAGE sample buffer, and immediately followed by an iced 1X Native-PAGE running buffer at 90 Volts for 3 hrs in a 4ºC circulating water bath. The proteins were stained and destained in the same way as for SDS-PAGE analysis.. 18.
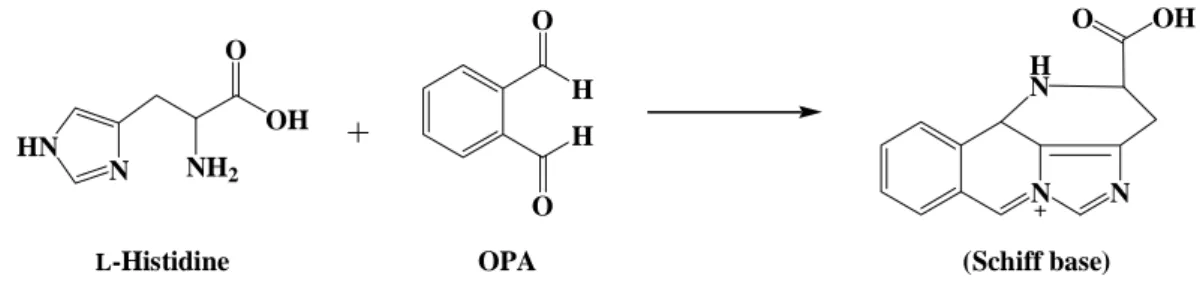
(35) Table 1: Solutions and volumes for the preparation of the SDS-PAGE and Native-PAGE separating gel and stacking gel. Separating gel. Stacking gel. 7.5%. 10%. 12%. 12.5%. 15%. 20%. 4%. ddH2O (mL). 16.8. 13.88. 11.55. 11. 8.05. 2.22. 1.7. 1.5 M Tris-HCl, pH 8.8 (mL). 8.75. 8.75. 8.75. 8.75. 8.75. 8.75. -. -. -. -. -. -. -. 1.25. 0.35. 0.35. 0.35. 0.35. 0.35. 0.35. 0.1. 8.75. 11.67. 14. 14.6. 17.5. 23.33. 0.01. 0.028. 0.014. 0.014. 0.014. 0.014. 0.014. 6.8. 0.35. 0.35. 0.35. 0.35. 0.35. 0.35. 10. 35. 35. 35. 35. 35. 35. 10. 1 M Tris-HCl, pH 6.8 (mL) 10 % SDS (mL)a 30% acrylamide / 1% N,N’-methylenediacrylamide (mL) TEMED (mL) 10 % Ammonium persulfate (APS)b (mL) Total (mL) a b. Replaced SDS with ddH2O while preparing Native-PAGE Recommended that it be prepared fresh and mixed at the end. 2.5 Enzymatic activity assay of PepD PepD activity was determined according to Teufel et al.[33] via the measurement of histidine, using an o-phthalaldehyde [16] reagent. The substrate, L-carnosine (β-Ala-L-His), was hydrolyzed to β-Alanine and L-Histidine. The fluorescence of the derivative of histidine with OPA was detected at λEx: 355 nm and λEm: 460 nm ( Fig. 6 ).. H2N O. H N. N. O. NH. OH. HO H. O. O. O. PepD H. β -Alanine. L-Carnosine. 19. NH2. OH HN. N. NH2. L-Histidine.
(36) H OH HN. N. H N. H. NH2. N. O L-Histidine. OH. O. O O. OPA. N. (Schiff base). Fig. 6: Formation of a Schiff base by L-histidine and o-phthalaldehyde. 20 μL purified enzyme (0.5 mg/mL) and 80 μL 50 mM Tris-HCl pH 6.8 buffer reacted with 0.5 mM L-carnosine over 20 min. Liberated histidine was derivatived by adding 100 μL OPA reagent, and incubated at 37ºC in darkness for 5 min. The reactions containing just buffer with L-histidine or L-carnosine reacting with OPA served as positive and negative controls, respectively. All reactions were carried out in triplicate. The fluorescence of the histidine derivatived with OPA was measured by Fluoroskan Ascent FL. (λEx: 355 nm and λEm: 460 nm).. 2.6 Mutagenesis analysis of V. alginolyticus pepD Site-directed mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Appendix 2) to create the mutants. Mutagenic primers were designed and pET-28a(+)-pepD plasmids (wild-type) were used as the template: the PCR reaction was carried out via the nonstrand-displacing action of pfuTurbo DNA polymerase to extend and incorporate the mutagenic primers (Appendix 3), resulting in the nicked circular strands. The PCR mutagenesis reaction was performed in a 96-well GeneAmp® PCR System 9700 Thermal Cycler, as recommended by the manufacturer of PfuUltraTM High-Fidelity DNA polymerase. For each reaction, 100 ng of wild-type plasmid, 5 μL 10X Pfu polymerase buffer, 4 μL 2.5 mM dNTP mix, 1 μL of each 12.5 μM primer, 1 μL (2.5 U) Pfu polymerase and ddH2O were added to the final volume of 50 μL (Table 3). The PCR products with wild-type 20.
(37) and mutant plasmids were incubated with DpnI for 4 hrs at 37ºC to selectively digest the methylated, non-mutated parental wild-type plasmids. After DpnⅠdigestion, the mutant plasmid was transformed into E. coli XL1-Blue competent cells, with selection for kanamycin resistance. After successful mutagenesis, which was confirmed by restriction enzymes and DNA sequencing of plasmid, the desired mutant plasmids were transformed into E. coli BL21( DE3 ) pLysS competent cells for expression of the mutant pepD proteins. Table 2: Reaction conditions and cycling parameters for the PCR mutagenesis reaction.. 2.7 The His-tag-cleaved PepD After purification of expressed Vibrio alginolyticus wild-type PepD (see 2.2), containing a His-tag at the N-terminal, the eluted PepD-containing protein fractions were incubated with thrombin (Sigma) in a cleavage buffer (40 mM Tris (pH 8.0), 300 mM NaCl, 2 mM CaCl2, and 5% glycerol) for 16 hours to digest the His-tag. After being subjected to benzamidine sepharoseTM6B (Pharmacia) to remove thrombin, the flow-through fractions were applied to another Ni-NTA column to collect and remove the His-tag fragments. The pooled native-form PepD proteins then were dialyzed with a 20mM HEPES buffer (pH 7.0), and stored at -80°C.. 21.
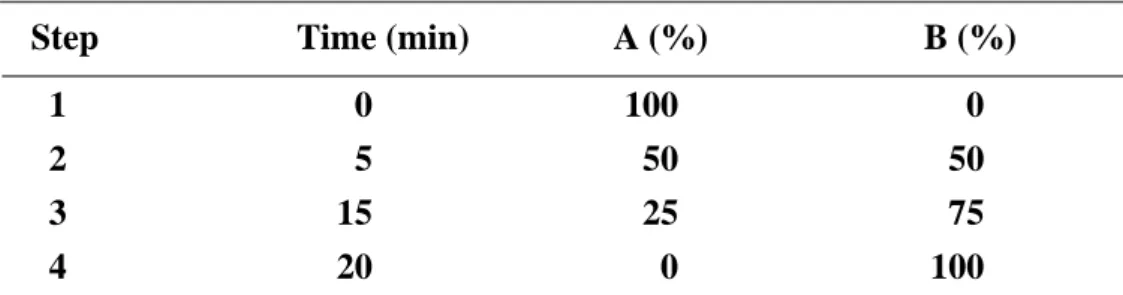
(38) 2.8 Metal ion effect of PepD activity The His-tag-cleaved PepD protein first was dialyzed overnight with buffer containing 20 mM MES pH6.0 and 5 mM EDTA to remove divalent zinc ion (apo-Pepd). The apo-PepD was dialyzed twice with 20 mM MES pH6.0 and exchanged with 20 mM HEPES pH7.0 before adding various divalent metal ions. The apo-PepD protein concentration was adjusted to 0.01 mM. Metal ions - including MgCl2, MnCl2, FeCl2, CoCl2, NiCl2, CuCl2, and CdCl2 were added into the final 20 mM HEPES buffer for metal exchange dialysis.. 2.9 Enzyme kinetics For determination of Vmax, Km, and kcat of the V. alginolyticus PepD, and to compare hydrolysis efficiency with the wild-type and mutant PepD, the method described by Csámpai et al.[47] was modified using High Performance Liquid Chromatography (HPLC) with a Fluorescence Detector (FLD). A system consisting of an Agilent 1100 Series Quaternary pump, Autosampler, Fluorescence Detector and Inertsil ODS-3 (7 μm, 7.6 mm×250 mm) column was used. The eluent system consisted of two components: eluent A was 0.05 M sodium acetate at pH 7.2, while eluent B was prepared from 0.1 M sodium acetate–acetonitrile–methanol (46:44:10, v/v/v) (titrated with glacial acetic acid or 1 M sodium hydroxide to pH 7.2). The gradient program was as described in Table 3. The fluent flow-rate was 0.8 mL/min at 30 ºC. Table 3: The fluent gradient program Step 1 2 3 4. Time (min) 0 5 15 20. A (%) 100 50 25 0. 22. B (%) 0 50 75 100.
(39) Different concentrations of L-carnosine (2, 1, 0.5, 0.25, 0.1 and 0.025 mM) were added as substrates to initiate enzymatic reactions. After 20 min incubation at 37 ºC, the samples were mixed with OPA reagent for 5 min incubation at 37ºC, then injected by autosampler. Fluorescence of the histidine with derivatived OPA was measured by FLD (λExc: 355 nm and λEm: 460 nm). Various concentrations of L-histidine solution (0.1, 0.05, 0.025, 0.01, 0.005, and 0.0025 mM) derivatived with OPA reagent were detected, using the method described above, to serve as standards.. 23.
(40) Chapter 3 Results. 3.1 Expression and Purification of Vibrio alginolyticus PepD On the basis of previous investigations in our laboratory, the V. alginolyticus pepD gene was sub-cloned into the expression plasmid pET28a(+), as a fragment that contained 1473 nucleotides and coded for a polypeptide of 490 amino acids (Appendix 4). This then was transferred into E. coli BL21(DE3)(pLysS) cells to generate pET28a(+)-pepD recombinant plasmid to protein, expressed as a (His)6-PepD fusion protein for purification by Ni-NTA column chromatography [48]. The calculated molecular weight indicated that PepD preferred to form a homodimer in its native state, as determined by analytical sedimentation velocity ultracentrifugation (Appendix 6); and PepD demonstrated optimal activity at a pH between 6.8 and 7.4. The Ni-NTA resin-bound PepD was eluted to high purity by 20 mM Tris-HCl, 0.5 M NaCl, pH 6.8 buffer containing 200 mM imidazole. The purified fractions were collected and dialyzed with 20 mM Tris-HCl, pH 6.8 buffer at 4ºC to remove the salts. The protein dialyzed by SDS-PAGE was found to be a single band, approximately 54 kDa in molecular mass, quite close to the calculated molecular mass, 53.6 kDa. Consequently, the results obtained here were in close accordance with the references, though further investigations continued to be warranted to explore unknown components of this project.. 24.
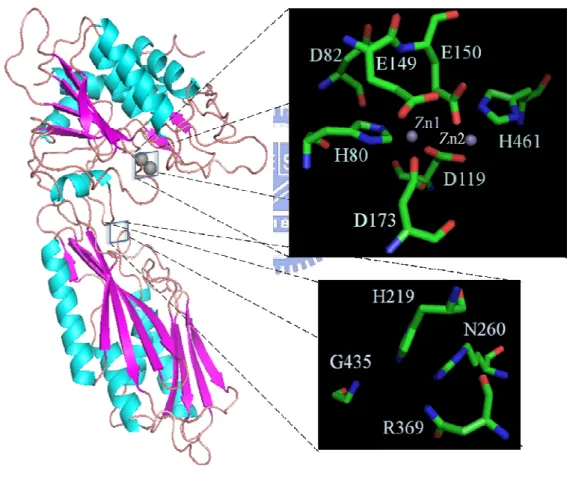
(41) 3.2 Multiple sequence alignment and structural model The structural model revealed that the side-chain ligands are disposed with two zinc ions (Zn1 and Zn2). The Zn1 is coordinated by the Nε2 of His80, one of the carboxylate oxygens of the bridging Asp119, and the carboxylate oxygen of Asp173. This model also demonstrates how Zn2 is coordinated with the other carboxylate oxygen of the bridging Asp119, the carboxylate oxygen of Glu150, and the His461 Nε2. (Fig. 7). Fig. 7: The overall structure of PepD and the local view of catalytic and zinc binding residues (by Chin-Yuan Chang). Five residues - His80, Asp119, Glu150, Asp173 and His461 - were felt to hold the catalytic metal ions. Furthermore, the structural model indicates that H219, N260, R369, and G435 are located in an important position. The bottom of overall structure was named lid domain and the part of top was catalytic domain. 25.
(42) In addition, a ‘bridging’ water molecule is predicted to exist between both zinc ions and close to the carboxylate group of the catalytic Glu150. Furthermore, the imidazole Nδ1 atoms of His80 (zinc 1) is further clamped to Asp82 O. In dinuclear zinc peptidases, the two catalytic zinc ions seem to have different tasks. Zn1 seems primarily to activate the catalytic water molecule and to promote binding and hydrolysis. On the other hand, Zn2 primarily appears to facilitate substrate binding and, with His219 Nε2, to polarize the scissile carbonyl group, thereby promoting nucleophilic attack by the catalytic water molecule.. Moreover, the substrate binding ability also was very important for these kinds of peptidase. Accordingly, to examine the overall structural features and the spatial locations, presuming that the substrate binding site of the V. alginolyticus PepD was obtained using PepV as the template, His219, Asn260, Arg369, and Gly435 were proposed as the putative substrate binding residues. (Fig. 8). Fig. 8: Local view of PepD (green) superimposed upon the substrate binding site of PepV (blue). The blue rods are residues of PepV, and the green rods are residues of PepD. AspΨ[PO2CH2]AlaOH is the phosphinate inhibitor. 26.
(43) The AspΨ[PO2CH2]AlaOH inhibitor mimics an sp3 hybridized peptide nitrogen of a bound dipeptide (Fig. 8). The C-terminal carboxylate group of the inhibitor extends into the ‘carboxylate groove’, where it is tightly fixed, via a network of hydrogen bonds, to surrounding polar groups. Furthermore, the environment around the C terminus of the bound dipeptide mimetic of PepD seems to be adapted to fix a dipeptide, with the carboxylate group framed by an extended hydrogen bond network, and the central Arg369 side chain and the Asn260 Nδ provided by the ‘lid domain’. The lid domain of PepD is important among the exopeptidases with respect to size and arrangement relative to the catalytic domain. Therefore, the lid domain of PepD is uniquely involved in substrate specificity.. Based upon the results of sequence alignment (Appendix 1) and structural model, five residues - including His80, Asp119, Glu150, Asp173 and His461 - appeared to hold the catalytic metal ions. Moreover, the - Asp82, Glu149, His219, Asn260, Arg369, and Gly435 were the putative catalysis and substrate binding residues (Fig. 9.). In order to understand these important functional residues, site-directed mutagenesis analysis and functional characterization of PepD with different metal ions were performed, as follows:. 27.
(44) Fig. 9: Nucleotide and predicted amino acid sequences of the V. alginolyticus pepD gene Proposed catalysis and substrate binding residues: Asp82, Glu149, His219, Asn260, Arg369, and His435. Proposed metal ion binding residues: His80, Asp119, Glu150, Asp173, His461. Proposed dimerization: Ile318-Ser397. 28.
(45) 3.3 Site-directed Mutagenesis Analysis of Vibrio alginolyticus PepD The function of Asp82 superimposed upon PepD is similar to that of Asp89 upon PepV. Asp82 is conserved in all the active enzymes of clan MH, considered to clamp the imidazolium ring of His80. Furthermore, the Nε2 of His80 is a coordinate of Zn1, and the Nδ1 is clamped to Asp82; besides, Glu150 is direct concerned with the metal binding. As a result, we wanted to mutate Asp82 and Glu150 of PepD by site-directed mutagenesis to learn more about the functional residues importance.. The mutants were generated using a QuikChange site-directed mutagenesis kit, as described in section 2.6, and the mutant plasmids transformed into E. coli. BL21(DE3)pLysS to express the mutant proteins. Following the same purification procedure as for V. alginolyticus wild-type PepD, the mutant PepD proteins were extracted using 20 mM Tris-HCl pH 6.8 buffer, containing 200 mM imidazole, by Ni-NTA column chromtography. The purified wild-type and mutant PepD proteins exhibited the same molecular weight, of about 55 kDa, on SDS-PAGE (Fig. 10). M. 1. 2. 3. 4. 5. 6. 7. 8. 9. 97 66 45 30 (kDa). Fig. 10: SDS-PAGE (12%) of purified wild-type and mutant proteins of Asp82 and Glu150. 29.
(46) Lane M: LMW protein marker; Lane 1: PepD wild type; Lane 2: PepD E150D mutant; Lane 3: PepD E150R mutant; Lane 4: PepD E150H mutant; Lane 5: PepD D82F mutant; Lane 6: PepD D82V mutant; Lane 7: PepD D82G mutant; Lane 8: PepD D82H; Lane 9: PepD D82E. The activity assays of the purified wild-type and mutant PepD proteins, taking L-carnosine as a substrate, were assessed here. The wild-type PepD catalyzed the hydrolysis. of L-carnosine under standard conditions, as described in section 2.5, its activity defined as 100% (Fig. 11).. Fig. 11: Enzymatic activities of wild-type and mutant Asp82 and Glu150 PepD on L-carnosine. The activity assay of the purified wild-type and mutant PepD proteins, taking L-carnosine as a substrate, were assessed. Wild-type activity was defined as 100%.. Asp82 was substituted with Gly, Val, Phe, Tyr, His, and Glu. As expected, no activity was detected for any of the Asp82 mutants. The substitution of Glu150 with Asp retained about 70% of the maximal hydrolytic activity of the wild-type enzyme, whereas substitution of Glu150 with Arg or His completely abolished enzymatic activity. 30.
(47) Moreover, based upon the structure model, His219, Asn260, Arg369, and Gly435 were identified as probable substrate binding residues, which may influence the catalytic mechanisms behind hydrolysis. Therefore, we created mutant PepD for these four residues, which were investigated using alanine scanning mutagenesis (Fig. 12). Arg369Ala lost enzymatic activity for hydrolyzing L-carnosine; but the other three residues did not relinquish catalytic activity, as predicted (Fig. 13). Accordingly, we calculated enzyme kinetics of these mutant proteins to determine the Vmax, Km and kcat values as compared with the wild-type PepD (Fig. 14)(Table 4).. Fig. 12: SDS-PAGE (12%) of purified wild-type and mutant proteins of His219, Asn260, Arg369, and Gly435. Lane M: LMW protein marker; Lane 1: PepD His219Ala mutant; Lane 2: PepD Asn260Ala mutant; Lane 3: PepD Arg369Ala mutant; Lane 4: PepD Gly435Ala; Lane 5-8: Western blot analysis of purified PepD mutanted protein with anti-PepD mAbs.. 31.
(48) Fig. 13: Enzymatic activities of wild-type and mutant His219, Asn260, Arg369, and Gly435 PepD on L-carnosine. The activity assay of the purified wild-type and mutant PepD proteins, taking L-carnosine as a substrate, were assessed here. Wild-type activity was defined as 100%.. Fig. 14: Enzyme kinetics of the mutant His219, Asn260, and Gly435 PepD proteins A Lineweaver-Burk plot, calculated from the respective Michaelis-Menten plot for mutant PepD proteins, demonstrated mutant PepD as a catalyst for the hydrolysis of 32.
(49) L-carnosine. Protein concentration of PepD was 1 uM, with catalytic reagents that catalyzed. the hydrolysis of L-carnosine in 50 mM Tris-HCl, pH 6.8 at 37oC.. Earlier investigations to determine kinetic has shown the apparent KM value of wild-type V. alginolyticus PepD activity on L-carnosine to be 0.36 mM. The turnover number (kcat) and catalytic efficiency (kcat/KM) of V. alginolyticus PepD were 8.6 min-1 and 0.398 mM-1s-1, respectively. The mutant PepD proteins were causing the various values shifted on kinetic parameters.. PepD. Kcat(min-1). Km(mM). Kcat/Km (mM-1s-1). WT. 8.6. 0.36. 0.398. H219A. 8. 0.24. 0.556. N260A. 6.94. 0.89. 0.130. G435A. 4.59. 0.28. 0.273. Table 4: Kinetic parameters for the hydrolysis of L-carnosine using mutant V.alginolyticus PepD.. 33.
(50) 3.4 Metal ion effect of PepD activity Several metallopeptidases are known to be key players in carcinogenesis, tissue repair, neurological processes, protein maturation, hormone-level regulation, cell-cycle control and protein-degradation. The majority of co-catalytic metallohydrolases are Zn2+-dependent enzymes, but some require other divalent metal ion. To chang metal ion may provide information about what possible role in enzyme function. In addition, aminoacyl-histidine dipeptidase (PepD) is a 54kD metallopeptidase, which is activated by Zn2+ as its wild-type.. The effect of metal substitution of the Vibrio alginolyticus PepD has been investigated and described in sections 2.2, 2.7, and 2.8. The apo-PepD was identified using a His-tag-cleaved and dialyzed buffer containing EDTA to remove divalent metal ions and yield the inactive protein. All the metal ions tested, including Mg2+, Mn2+, Co2+, Ni2+, Cu2+ and Cd2+, can activate apo-PepD, and exert some level of hydrolysis activity on L-carnosine. The different metal-substituted derivatives of PepD exhibit different levels of activity (Fig. 15).. Fig. 15: Metal ion effect on PepD activity. The activity assays were performed at 37°C for 30 min in the presence of 20 mM HEPES buffer, pH 7.0, 2 mM L-carnosine, 10 μM of 34.
(51) purified enzyme, and 20 μM of the different metal salts. The activity was measured according to standard activity assay protocol. Values are expressed as relative activity, based upon setting the hydrolysis of L-carnosine at 100%.. Particularly, the Mn2+, Co2+, Ni2+, Cu2+ and Cd2+ substituted derivatives of PepD exhibited higher levels of activity than the native PepD, in terms of the hydrolysis of 2+. L-carnosine. Conversely, no enzymatic activity was detected with Fe , and the substitution. of Zn2+ with Mg2+ resulted in ~70% restoration of optimal enzymatic activity.. 35.
(52) 3.5 Enzyme kinetics of the metal effects on Vibrio alginolyticus PepD Interestingly, different metal ions produced different levels of enzymatic activity of V. alginolyticus PepD in the hydrolysis of L-carnosine. Therefore, we investigated the enzyme kinetics of V. alginolyticus PepD for L-carnosine in greated depth, as described in section 2.9 (Fig. 16).. Fig. 16: Enzyme kinetics of the metal effects on Vibrio alginolyticus PepD A Lineweaver-Burk plot, calculated from the respective Michaelis-Menten plot for metal ions, demonstrated metal ion effects on PepD as a catalyst for the hydrolysis of L-carnosine. Protein concentration of PepD was 1 uM, with catalytic reagents that catalyzed the hydrolysis of L-carnosine in 50 mM Tris-HCl, pH 6.8 at 37oC.. Furthermore, the enzyme kinetics of the PepD complex with Co2+, Ni2+, and Cd2+ for hydrolyzing L-carnosine were determined as Km and kcat (Fig. 16). The turnover number (kcat, 36.
(53) kcat = Vmax/[E]t ) and catalytic efficiency (kcat/ Km) of Co, Ni, and Cd substituted derivatives of PepD were 23.3, 12.8, 23.7 (min-1) and 1.24, 1.01, 1.10 (mM-1s-1), respectively (Table 5).. PepD. Kcat(min-1). Km(mM). Kcat/Km (mM-1s-1). WT. 8.6. 0.36. 0.398. Co-WT. 23.3. 0.31. 1.24. Ni-WT. 12.8. 0.21. 1.01. Cd-WT. 23.7. 0.36. 1.10. Table 5: Kinetic parameters for the hydrolysis of L-carnosine using different metal ion derivatives that complex with V. alginolyticus PepD.. 37.
(54) Chapter 4 Discussion and Conclusions V. alginolyticus PepD, which has been identified as a member of the metallopeptidase M20 family and is considered an aminoacylhistidine dipeptidase, was investigated. The putative active site of V. alginolyticus PepD could be proposed on the basis of sequence analysis and from the modeling results. We have successfully expressed and purified wild-type PepD and mutants from V. alginolyticus. The way V. alginolyticus PepD hydrolyzes L-carnosine (β-Ala-L-His) is similar to how other known PepD do this.. Asp82 is two residues downstream from His80, in the vicinity of the zinc center, and is thought to clamp the imidazolium ring of His80 Nδ1. Additionally, potential hydrogen-bonding interactions of the Nδ1 porton of His80 with a side chain oxygen of Asp82 forming an Asp-His-Zn1 triad that has been postulated to decrease the Lewis acidity of Zn2+ and may further assist in facilitating the coordination of a double-bonded oxygen to Zn1(Fig.17 step 0 - 2). A catalytic mechanism is proposed that the bridging catalytic water attacks the carbonyl carbon of the scissile peptide bond to form a sp3-orbital substrate-enzyme tetrahedral intermediate (Fig.17 step 3 - 4). Therefore, not only does the Zn2 coordinated by the carboxylate oxygen of Glu150 cause these Glu150 mutants to lose enzyme activity, but the Asp82 mutants also experience lost enzyme activity, suggesting that the probable role of Asp82 on PepD is clamping the imidazolium ring of His80 Nδ1, so as to correctly coordinate Zn1 to enhance substrate and metal binding ability. The substitution of Glu150 to Asp was resulted in only partial loss of the enzymatic activity; maybe the replacement of Glu with Asp at this position only partially affects the metal ligand-binding affinity and subsequent activation of the catalytic water for substrate-enzyme tetrahedral intermediate formation.. 38.
(55) Fig. 17: Proposed mechanism for the hydrolysis of PepD, catalyzed by a metallopeptidase with a co-catalytic active site, where R1, R2 and R3 are substrate side chains, and R is an N-terminal amine or a C-terminal carboxylate. This mechanism is based upon the proposed mechanism for the aminopeptidase from Aeromonas proteolytica.. On the basis of structural model superposition of PepD on PepV after optimal fit, the substrate binding site residues and the metal center catalytic domain both are shown to be important. After mutation analysis of several residues, only Arg369 exhibited reduced enzymatic activity to hydrolyze L-carnosine as a substrate. The role of Arg369 not only is located at the probable dimerization domain, but it also appears to be that part of the lid domain, at which the substrate carboxyl end should be trapped in the hydrogen bond network dominated by the Arg369 guanidyl group, that contributes to the tightening of substrate 39.
(56) binding (Fig.17 step 1). Unfortunately, the mutant PepD of the other three residues did not lose all of catalytic activity, as expeced; and that His219Ala increased enzymatic activity was a surprise. The enzyme kinetics study showed that the relative activities of His219Ala mutant protein in terms of kcat and kcat/Km were better then wild-type. Moreover, the Km value of Asn260Ala suggested the mutant of this residue was influencing on substrate binding ability, and the shifted kcat value of Gly435Ala indicate that was influencing on catalytic activity. Maybe the key conclusions that can be drawn from this study are that these residues need to be studied in greater depth.. V. alginolyticus PepD is a 54kD metallopeptidase, which is activated by Zn2+ in its wild-type. The functional importance of metal ions - including Co2+, Cu2+, Ni2+, Mg2+, Mn2+ and Cd2+ - may be indicated by the different levels of restored activity and the apparent value shift to kcat in enzyme kinetics of the reconstituted metal PepD protein. Possible roles for both metal ions in PepD that contains co-catalytic active sites include: (i) binding and positioning substrate; (ii) binding and activating a water molecule to yield an active site hydroxide nucleophile; and (iii) stabilizing the transition state of a hydrolytic reaction. The Lewis acidity of the metal ions is the key factor in metal-centered hydrolysis, in which the pKa of the coordinated water is significantly lowered to assist its nucleophilic attack on the scissile bond at a neutral pH. Consistent with this hypothesis, both Mg2+ and Mn2+ ions bind to the active site of APPro in a very similar resule, which Mn2+ activates APPro and Mg2+ does not [49]. Mg2+ has a higher charge density and is a harder Lewis acid than any other metal ion that is widely available in biological systems. This property makes Mg2+ the perfect metal ion for binding to the hard oxygen anions of the negatively-charged phosphodiester backbone of nucleic acids. As a result, Mg2+ is a good key factor for nucleases and other phosphohydrolases, but not for peptidase. Zn2+, Mn2+ and Co2+ have a lower charge density than Mg2+, and are stronger Lewis acids, based upon their respective ionization potentials. 40.
(57) Perhaps that is the reason for nature’s apparent choice of Zn2+, Mn2+ or Co2+ at the catalytically-active site for most peptidases.. In conclusion, V. alginolyticus PepD is an aminoacylhistidine dipeptidase that can hydrolyze Xaa-His dipeptides, including an unusual dipeptide - carnosine (β-Ala-L-His). To confirm this, further investigations are warranted into the putative importance of functional residues that almost exhibit a decrease or lose of enzymatic activity. Our results reveal that these residues are involved in both substrate and metal binding, and that this dramatically affects enzymatic activity. The addition of metal ions led to the alteration in the specific activity of recombinant PepD and the activity is increased by metal ions that complex with PepD , like Mn2+, Co2+, Ni2+ and Cd2+. The improvement of activity and stability by modification of metal species would be promising from an aspect of the practical application of the enzyme.. 41.
(58) Chapter 5 Future work Placing the structural model of Vibrio alginolyticus PepD at the proposed substrate binding site reveals that those residues create a place like a ‘carboxylate groove’ to tightly fix the substrate at the lid domain, via a network of hydrogen bonds. We identified several important residues, among which Arg369Ala loses enzymatic activity and His219Ala has it increased, that should be investigated further by site-directed mutagenesis with other amino acids and the ducking method. Moreover, Asp173 was present in homologues with aminopeptidase/dipeptidase. specificity,. whereas. members. of. the. aminoacylase/carboxypeptidase family possessed a glutamic acid at the same position. Therefore, this residue also could be investigated by site-directed mutagenesis.. The mechanisms for hydrolyses of phosphoesters and peptides are quite different, but recent studies have shown that different metal ions at the catalytic center where the peptidase changes are substrate specific, and these metal-substituted derivatives of peptidase could hydrolyze phosphate ester bonds. We produced several metal-substituted PepD that could be further investigated for substrate specificity, especially those that hydrolyze phosphoesters as a substrate. Having the necessary structure in neither the peptidase family M20 nor similar peptidases, crystallization of the V. alginolyticus PepD was needed. Furthermore, the crystalline structure of the mutant PepD proteins combined with the wild-type structure, and mutagenesis analysis data could provide insights into the catalytic mechanism of bacterial aminoacylhistidine dipeptidase.. 42.
(59) Chapter 6 Reference [1]. [2] [3] [4]. [5]. [6] [7]. [8]. [9]. [10]. [11] [12]. [13] [14]. Goarant, C., Merien, F., Berthe, F., Mermoud, I. and Perolat, P. (1999). Arbitrarily primed PCR to type Vibrio spp. pathogenic for shrimp. Appl Environ Microbiol 65, 1145-51. Miyamoto, Y., Nakamura, K., and Takizawa, K. (1961). Proposals of a new genus “Oceanomonas” and of the amended species names. Jpn. J. Microbiol. 5, 477-86 Sakazaki, R. (1968). Proposal of Vibrio alginolyticus for the biotype 2 of Vibrio parahaemolyticus. Jpn J Med Sci Biol 21, 359-62. Molitoris, E., Joseph, S.W., Krichevsky, M.I., Sindhuhardja, W. and Colwell, R.R. (1985). Characterization and distribution of Vibrio alginolyticus and Vibrio parahaemolyticus isolated in Indonesia. Appl Environ Microbiol 50, 1388-94. Baross, J. and Liston, J. (1970). Occurrence of Vibrio parahaemolyticus and related hemolytic vibrios in marine environments of Washington State. Appl Microbiol 20, 179-86. Lee, K.K. (1995). Pathogenesis studies on Vibrio alginolyticus in the grouper, Epinephelus malabaricus, Bloch et Schneider. Microb Pathog 19, 39-48. Licciano, M., Stabili, L. and Giangrande, A. (2005). Clearance rates of Sabella spallanzanii and Branchiomma luctuosum (Annelida: Polychaeta) on a pure culture of Vibrio alginolyticus. Water Res 39, 4375-84. Lee, K.K., Yu, S.R., Chen, F.R., Yang, T.I. and Liu, P.C. (1996). Virulence of Vibrio alginolyticus isolated from diseased tiger prawn, Penaeus monodon. Curr Microbiol 32, 229-31. Zen-Yoji, H., Le Clair, R.A., Ota, K. and Montague, T.S. (1973). Comparison of Vibrio parahaemolyticus cultures isolated in the United States with those isolated in Japan. J Infect Dis 127, 237-41. Levine, W.C. and Griffin, P.M. (1993). Vibrio infections on the Gulf Coast: results of first year of regional surveillance. Gulf Coast Vibrio Working Group. J Infect Dis 167, 479-83. Reina Prieto, J. and Hervas Palazon, J. (1993). Otitis media due to Vibrio alginolyticus: the risks of the Mediterranean Sea. An Esp Pediatr 39, 361-3. Gahrn-Hansen, B. and Hornstrup, M.K. (1994). Extraintestinal infections caused by Vibrio parahaemolyticus and Vibrio alginolyticus at the county of Funen 1987-1992. Ugeskr Laeger 156, 5279-82. Rippey, S.R. (1994). Infectious diseases associated with molluscan shellfish consumption. Clin Microbiol Rev 7, 419-25. Rose, J.B., Epstein, P.R., Lipp, E.K., Sherman, B.H., Bernard, S.M. and Patz, J.A. 43.
(60) [15] [16] [17] [18] [19] [20] [21]. [22]. [23] [24]. [25] [26] [27] [28]. [29]. (2001). Climate variability and change in the United States: potential impacts on water- and foodborne diseases caused by microbiologic agents. Environ Health Perspect 109 Suppl 2, 211-21. Hlady, W.G. and Klontz, K.C. (1996). The epidemiology of Vibrio infections in Florida, 1981-1993. J Infect Dis 173, 1176-83. Opal, S.M. and Saxon, J.R. (1986). Intracranial infection by Vibrio alginolyticus following injury in salt water. J Clin Microbiol 23, 373-4. Matsiota-Bernard, P. and Nauciel, C. (1993). Vibrio alginolyticus wound infection after exposure to sea water in an air crash. Eur J Clin Microbiol Infect Dis 12, 474-5. Wilcox, D.E. (1996). Binuclear Metallohydrolases. Chem Rev 96, 2435-2458. Dismukes, G.C. (1996). Manganese Enzymes with Binuclear Active Sites. Chem Rev 96, 2909-2926. Lipscomb, W.N. and Strater, N. (1996). Recent Advances in Zinc Enzymology. Chem Rev 96, 2375-2434. Munih, P., Moulin, A., Stamper, C.C., Bennett, B., Ringe, D., Petsko, G.A. and Holz, R.C. (2007). X-ray crystallographic characterization of the Co(II)-substituted Tris-bound form of the aminopeptidase from Aeromonas proteolytica. J Inorg Biochem 101, 1099-107. Chen, S.L., Marino, T., Fang, W.H., Russo, N. and Himo, F. (2008). Peptide hydrolysis by the binuclear zinc enzyme aminopeptidase from Aeromonas proteolytica: a density functional theory study. J Phys Chem B 112, 2494-500. Rawlings, N.D. and Barrett, A.J. (1993). Evolutionary families of peptidases. Biochem J 290 ( Pt 1), 205-18. Lindner, H.A., Alary, A., Boju, L.I., Sulea, T. and Menard, R. (2005). Roles of dimerization domain residues in binding and catalysis by aminoacylase-1. Biochemistry 44, 15645-51. Rawlings, N.D., Morton, F.R. and Barrett, A.J. (2006). MEROPS: the peptidase database. Nucleic Acids Res 34, D270-2. Rawlings, N.D. and Barrett, A.J. (1995). Evolutionary families of metallopeptidases. Methods Enzymol 248, 183-228. van der Drift, C. and Ketelaars, H.C. (1974). Carnosinase: its presence in Pseudomonas aeruginosa. Antonie Van Leeuwenhoek 40, 377-84. Brombacher, E., Dorel, C., Zehnder, A.J. and Landini, P. (2003). The curli biosynthesis regulator CsgD co-ordinates the expression of both positive and negative determinants for biofilm formation in Escherichia coli. Microbiology 149, 2847-57. Schroeder, U., Henrich, B., Fink, J. and Plapp, R. (1994). Peptidase D of Escherichia coli K-12, a metallopeptidase of low substrate specificity. FEMS Microbiol Lett 123, 153-9. 44.
數據
相關文件
SF15140A 楊勝舜 利用 JKB-122 評估對於經干擾素(長效 型或短效型)或干擾素和 Ribavirin 組合 治療沒有反應的 C 型肝炎病毒陽性患 者之肝臟功能 (丙胺酸轉胺酶
提高免疫力上有良好的功效。可治 療糖尿病血管併發症及扁平疣、傳染性疣。在動物 試驗中,利用環磷醯胺製出免
However, due to the multi-disciplinary nature of this subject, schools may consider assigning teachers with different expertise to teach this subject at different levels (S4, 5
◦ 金屬介電層 (inter-metal dielectric, IMD) 是介於兩 個金屬層中間,就像兩個導電的金屬或是兩條鄰 近的金屬線之間的絕緣薄膜,並以階梯覆蓋 (step
• Strange metal state are generic non-Fermi liquid properties in correlated electron systems near quantum phase transitions. • Kondo in competition with RVB spin-liquid provides
Basing on the observation and assessment results, this study analyzes and discusses the effects and problems of learning the polynomial derivatives on different level students
危機事件 後果 可預測性 持續性 震撼程度 估計危機 影響程度 一對小四及小. 二的兄妹,居 於學校同邨的
廢鹵化溶劑 易燃,有害 紫色 廢酸性液體,FeCl 3 ,蝕板液 腐蝕 黃色 廢酸性液體,HNO 3 ,蝕板液 剌激 橙色. 廢機油 易燃